Distinct modes of regulation by chromatin encoded through nucleosome positioning signals
- PMID: 18989395
- PMCID: PMC2570626
- DOI: 10.1371/journal.pcbi.1000216
Distinct modes of regulation by chromatin encoded through nucleosome positioning signals
Abstract
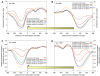
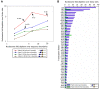
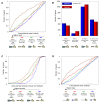
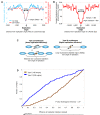
The detailed positions of nucleosomes profoundly impact gene regulation and are partly encoded by the genomic DNA sequence. However, less is known about the functional consequences of this encoding. Here, we address this question using a genome-wide map of approximately 380,000 yeast nucleosomes that we sequenced in their entirety. Utilizing the high resolution of our map, we refine our understanding of how nucleosome organizations are encoded by the DNA sequence and demonstrate that the genomic sequence is highly predictive of the in vivo nucleosome organization, even across new nucleosome-bound sequences that we isolated from fly and human. We find that Poly(dA:dT) tracts are an important component of these nucleosome positioning signals and that their nucleosome-disfavoring action results in large nucleosome depletion over them and over their flanking regions and enhances the accessibility of transcription factors to their cognate sites. Our results suggest that the yeast genome may utilize these nucleosome positioning signals to regulate gene expression with different transcriptional noise and activation kinetics and DNA replication with different origin efficiency. These distinct functions may be achieved by encoding both relatively closed (nucleosome-covered) chromatin organizations over some factor binding sites, where factors must compete with nucleosomes for DNA access, and relatively open (nucleosome-depleted) organizations over other factor sites, where factors bind without competition.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures





Similar articles
-
A genomic code for nucleosome positioning.Nature. 2006 Aug 17;442(7104):772-8. doi: 10.1038/nature04979. Epub 2006 Jul 19. Nature. 2006. PMID: 16862119 Free PMC article.
-
Nucleosome positioning in yeasts: methods, maps, and mechanisms.Chromosoma. 2015 Jun;124(2):131-51. doi: 10.1007/s00412-014-0501-x. Epub 2014 Dec 23. Chromosoma. 2015. PMID: 25529773 Review.
-
Z curve theory-based analysis of the dynamic nature of nucleosome positioning in Saccharomyces cerevisiae.Gene. 2013 Nov 1;530(1):8-18. doi: 10.1016/j.gene.2013.08.018. Epub 2013 Aug 16. Gene. 2013. PMID: 23958656
-
Translational and rotational settings of H2A.Z nucleosomes across the Saccharomyces cerevisiae genome.Nature. 2007 Mar 29;446(7135):572-6. doi: 10.1038/nature05632. Nature. 2007. PMID: 17392789
-
Genome-Wide Analysis of Nucleosome Positions, Occupancy, and Accessibility in Yeast: Nucleosome Mapping, High-Resolution Histone ChIP, and NCAM.Curr Protoc Mol Biol. 2014 Oct 1;108:21.28.1-21.28.16. doi: 10.1002/0471142727.mb2128s108. Curr Protoc Mol Biol. 2014. PMID: 25271716 Free PMC article. Review.
Cited by
-
Distal chromatin structure influences local nucleosome positions and gene expression.Nucleic Acids Res. 2012 May;40(9):3870-85. doi: 10.1093/nar/gkr1311. Epub 2012 Jan 12. Nucleic Acids Res. 2012. PMID: 22241769 Free PMC article.
-
Defining flexible vs. inherent promoter architectures: the importance of dynamics and environmental considerations.Nucleus. 2012 Sep-Oct;3(5):399-403. doi: 10.4161/nucl.21172. Epub 2012 Jul 3. Nucleus. 2012. PMID: 22751015 Free PMC article.
-
H2A.Z nucleosome positioning has no impact on genetic variation in Drosophila genome.PLoS One. 2013;8(3):e58295. doi: 10.1371/journal.pone.0058295. Epub 2013 Mar 5. PLoS One. 2013. PMID: 23472174 Free PMC article.
-
Systematic dissection of roles for chromatin regulators in a yeast stress response.PLoS Biol. 2012;10(7):e1001369. doi: 10.1371/journal.pbio.1001369. Epub 2012 Jul 31. PLoS Biol. 2012. PMID: 22912562 Free PMC article.
-
Interplay between genetic, epigenetic, and gene expression variability: Considering complexity in evolvability.Evol Appl. 2021 Feb 19;14(4):893-901. doi: 10.1111/eva.13204. eCollection 2021 Apr. Evol Appl. 2021. PMID: 33897810 Free PMC article. Review.
References
-
- van Holde KE. Chromatin. New York: Springer; 1989.
-
- Ioshikhes IP, Albert I, Zanton SJ, Pugh BF. Nucleosome positions predicted through comparative genomics. Nat Genet. 2006;38:1210–1215. - PubMed
-
- Lee W, Tillo D, Bray N, Morse RH, Davis RW, et al. A high-resolution atlas of nucleosome occupancy in yeast. Nat Genet. 2007;39:1235–1244. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

