miR-296 regulates growth factor receptor overexpression in angiogenic endothelial cells
- PMID: 18977327
- PMCID: PMC2597164
- DOI: 10.1016/j.ccr.2008.10.005
miR-296 regulates growth factor receptor overexpression in angiogenic endothelial cells
Abstract
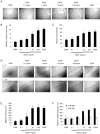
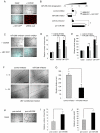
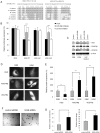
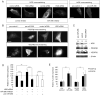
A key step in angiogenesis is the upregulation of growth factor receptors on endothelial cells. Here, we demonstrate that a small regulatory microRNA, miR-296, has a major role in this process. Glioma cells and angiogenic growth factors elevate the level of miR-296 in primary human brain microvascular endothelial cells in culture. The miR-296 level is also elevated in primary tumor endothelial cells isolated from human brain tumors compared to normal brain endothelial cells. Growth factor-induced miR-296 contributes significantly to angiogenesis by directly targeting the hepatocyte growth factor-regulated tyrosine kinase substrate (HGS) mRNA, leading to decreased levels of HGS and thereby reducing HGS-mediated degradation of the growth factor receptors VEGFR2 and PDGFRbeta. Furthermore, inhibition of miR-296 with antagomirs reduces angiogenesis in tumor xenografts in vivo.
Figures


Similar articles
-
Nuclear receptor COUP-TFII controls pancreatic islet tumor angiogenesis by regulating vascular endothelial growth factor/vascular endothelial growth factor receptor-2 signaling.Cancer Res. 2010 Nov 1;70(21):8812-21. doi: 10.1158/0008-5472.CAN-10-0551. Epub 2010 Oct 26. Cancer Res. 2010. PMID: 20978203 Free PMC article.
-
MiR-383 inhibits proliferation, migration and angiogenesis of glioma-exposed endothelial cells in vitro via VEGF-mediated FAK and Src signaling pathways.Cell Signal. 2017 Jan;30:142-153. doi: 10.1016/j.cellsig.2016.09.007. Epub 2016 Sep 28. Cell Signal. 2017. PMID: 27693218
-
Flt-1 regulates vascular endothelial cell migration via a protein tyrosine kinase-7-dependent pathway.Blood. 2011 May 26;117(21):5762-71. doi: 10.1182/blood-2010-09-306928. Epub 2011 Apr 1. Blood. 2011. PMID: 21460247 Free PMC article.
-
Angiogenesis-related growth factors in brain tumors.Cancer Treat Res. 2004;117:169-90. doi: 10.1007/978-1-4419-8871-3_12. Cancer Treat Res. 2004. PMID: 15015561 Review.
-
Growth factors in glioma angiogenesis: FGFs, PDGF, EGF, and TGFs.J Neurooncol. 2000 Oct-Nov;50(1-2):121-37. doi: 10.1023/a:1006436624862. J Neurooncol. 2000. PMID: 11245272 Review.
Cited by
-
Endocytosis of receptor tyrosine kinases.Cold Spring Harb Perspect Biol. 2013 May 1;5(5):a017459. doi: 10.1101/cshperspect.a017459. Cold Spring Harb Perspect Biol. 2013. PMID: 23637288 Free PMC article. Review.
-
MicroRNAs as diagnostic and prognostic biomarkers of age-related macular degeneration: advances and limitations.Neural Regen Res. 2021 Mar;16(3):440-447. doi: 10.4103/1673-5374.293131. Neural Regen Res. 2021. PMID: 32985463 Free PMC article. Review.
-
Recent advances in the molecular understanding of glioblastoma.J Neurooncol. 2012 May;108(1):11-27. doi: 10.1007/s11060-011-0793-0. Epub 2012 Jan 20. J Neurooncol. 2012. PMID: 22270850 Free PMC article. Review.
-
The role of microRNA in the pathogenesis of glial brain tumors.Noncoding RNA Res. 2022 Feb 25;7(2):71-76. doi: 10.1016/j.ncrna.2022.02.005. eCollection 2022 Jun. Noncoding RNA Res. 2022. PMID: 35330864 Free PMC article. Review.
-
MicroRNAs in vascular biology and vascular disease.J Cardiovasc Transl Res. 2010 Jun;3(3):235-40. doi: 10.1007/s12265-010-9164-z. Epub 2010 Feb 13. J Cardiovasc Transl Res. 2010. PMID: 20560045 Free PMC article. Review.
References
-
- Ambros V. The functions of animal microRNAs. Nature. 2004;431:350–355. - PubMed
-
- Bache KG, Raiborg C, Mehlum A, Stenmark H. STAM and Hrs are subunits of a multivalent ubiquitin-binding complex on early endosomes. J Biol Chem. 2003;278:12513–12521. - PubMed
-
- Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. - PubMed
-
- Brem S, Cotran R, Folkman J. Tumor angiogenesis: a quantitative method for histologic grading. J Natl Cancer Inst. 1972;48:347–356. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

