Design principles of biochemical oscillators
- PMID: 18971947
- PMCID: PMC2796343
- DOI: 10.1038/nrm2530
Design principles of biochemical oscillators
Abstract
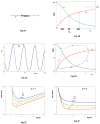
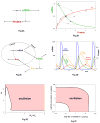
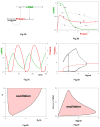
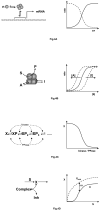
Cellular rhythms are generated by complex interactions among genes, proteins and metabolites. They are used to control every aspect of cell physiology, from signalling, motility and development to growth, division and death. We consider specific examples of oscillatory processes and discuss four general requirements for biochemical oscillations: negative feedback, time delay, sufficient 'nonlinearity' of the reaction kinetics and proper balancing of the timescales of opposing chemical reactions. Positive feedback is one mechanism to delay the negative-feedback signal. Biological oscillators can be classified according to the topology of the positive- and negative-feedback loops in the underlying regulatory mechanism.
Figures
Similar articles
-
Analytical study of robustness of a negative feedback oscillator by multiparameter sensitivity.BMC Syst Biol. 2014;8 Suppl 5(Suppl 5):S1. doi: 10.1186/1752-0509-8-S5-S1. Epub 2014 Dec 12. BMC Syst Biol. 2014. PMID: 25605374 Free PMC article.
-
Circadian clocks: self-assembling oscillators?Curr Biol. 2003 Sep 2;13(17):R681-2. doi: 10.1016/s0960-9822(03)00608-0. Curr Biol. 2003. PMID: 12956973 Review.
-
Design principles underlying circadian clocks.J R Soc Interface. 2004 Nov 22;1(1):119-30. doi: 10.1098/rsif.2004.0014. J R Soc Interface. 2004. PMID: 16849158 Free PMC article.
-
Homogeneous Time Constants Promote Oscillations in Negative Feedback Loops.ACS Synth Biol. 2018 Jun 15;7(6):1481-1487. doi: 10.1021/acssynbio.7b00442. Epub 2018 May 14. ACS Synth Biol. 2018. PMID: 29676894 Free PMC article.
-
Essential and expendable features of the circadian timekeeping mechanism.Curr Opin Neurobiol. 2006 Dec;16(6):686-92. doi: 10.1016/j.conb.2006.09.001. Epub 2006 Sep 29. Curr Opin Neurobiol. 2006. PMID: 17011182 Review.
Cited by
-
Beyond symmetry-breaking: competition and negative feedback in GTPase regulation.Trends Cell Biol. 2013 Oct;23(10):476-83. doi: 10.1016/j.tcb.2013.05.003. Epub 2013 May 31. Trends Cell Biol. 2013. PMID: 23731999 Free PMC article. Review.
-
Tuning genetic clocks employing DNA binding sites.PLoS One. 2012;7(7):e41019. doi: 10.1371/journal.pone.0041019. Epub 2012 Jul 31. PLoS One. 2012. PMID: 22859962 Free PMC article.
-
Identification of genes with oscillatory expression in glioblastoma: the paradigm of SOX2.Sci Rep. 2024 Jan 24;14(1):2123. doi: 10.1038/s41598-024-51340-z. Sci Rep. 2024. PMID: 38267500 Free PMC article.
-
A dynamic and intricate regulatory network determines Pseudomonas aeruginosa virulence.Nucleic Acids Res. 2013 Jan 7;41(1):1-20. doi: 10.1093/nar/gks1039. Epub 2012 Nov 11. Nucleic Acids Res. 2013. PMID: 23143271 Free PMC article. Review.
-
Is the diatom sex clock a clock?J R Soc Interface. 2021 Jun;18(179):20210146. doi: 10.1098/rsif.2021.0146. Epub 2021 Jun 16. J R Soc Interface. 2021. PMID: 34129790 Free PMC article.
References
-
- Hess B, Boiteux A. Oscillatory phenomena in biochemistry. Annu Rev Biochem. 1971;40:237–258. - PubMed
-
- Gerisch G, Fromm H, Huesgen A, Wick U. Control of cell-contact sites by cyclic AMP pulses in differentiating Dictyostelium cells. Nature. 1975;255:547–549. - PubMed
-
- Olsen LF, Degn H. Oscillatory kinetics of the peroxidase-oxidase reaction in an open system. Experimental and theoretical studies. Biochim Biophys Acta. 1978;523:321–334. - PubMed
-
- Olsen LF, Degn H. Chaos in an enzyme reaction. Nature. 1977;267:177–178. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

