An equivalent metal ion in one- and two-metal-ion catalysis
- PMID: 18953336
- PMCID: PMC2597392
- DOI: 10.1038/nsmb.1502
An equivalent metal ion in one- and two-metal-ion catalysis
Abstract
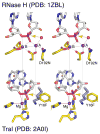
Nucleotidyl-transfer enzymes, which synthesize, degrade and rearrange DNA and RNA, often depend on metal ions for catalysis. All DNA and RNA polymerases, MutH-like or RNase H-like nucleases and recombinases, and group I introns seem to require two divalent cations to form a complete active site. The two-metal-ion mechanism has been proposed to orient the substrate, facilitate acid-base catalysis and allow catalytic specificity to exceed substrate binding specificity attributable to the stringent metal-ion (Mg2+ in particular) coordination. Not all nucleotidyl-transfer enzymes use two metal ions for catalysis, however. The betabetaalpha-Me and HUH nucleases depend on a single metal ion in the active site for the catalysis. All of these one- and two metal ion-dependent enzymes generate 5'-phosphate and 3'-OH products. Structural and mechanistic comparisons show that these seemingly unrelated nucleotidyl-transferases share a functionally equivalent metal ion.
Figures
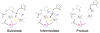
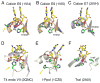
Similar articles
-
Metal-ion-dependent catalysis and specificity of CCA-adding enzymes: a comparison of two classes.Biochemistry. 2005 Sep 27;44(38):12849-59. doi: 10.1021/bi0509402. Biochemistry. 2005. PMID: 16171400
-
Crystal structures of RNase H bound to an RNA/DNA hybrid: substrate specificity and metal-dependent catalysis.Cell. 2005 Jul 1;121(7):1005-16. doi: 10.1016/j.cell.2005.04.024. Cell. 2005. PMID: 15989951
-
A combined experimental and theoretical study of divalent metal ion selectivity and function in proteins: application to E. coli ribonuclease H1.J Am Chem Soc. 2003 Aug 6;125(31):9318-28. doi: 10.1021/ja034956w. J Am Chem Soc. 2003. PMID: 12889961
-
Catalytic metal ions and enzymatic processing of DNA and RNA.Acc Chem Res. 2015 Feb 17;48(2):220-8. doi: 10.1021/ar500314j. Epub 2015 Jan 15. Acc Chem Res. 2015. PMID: 25590654 Review.
-
Multiple roles of metal ions in large ribozymes.Met Ions Life Sci. 2011;9:197-234. doi: 10.1039/9781849732512-00197. Met Ions Life Sci. 2011. PMID: 22010273 Review.
Cited by
-
Viewing Human DNA Polymerase β Faithfully and Unfaithfully Bypass an Oxidative Lesion by Time-Dependent Crystallography.J Am Chem Soc. 2015 Apr 22;137(15):5225-30. doi: 10.1021/jacs.5b02109. Epub 2015 Apr 9. J Am Chem Soc. 2015. PMID: 25825995 Free PMC article.
-
The CRISPR-associated DNA-cleaving enzyme Cpf1 also processes precursor CRISPR RNA.Nature. 2016 Apr 28;532(7600):517-21. doi: 10.1038/nature17945. Epub 2016 Apr 20. Nature. 2016. PMID: 27096362
-
Distal Mutations in the β-Clamp of DNA Polymerase III* Disrupt DNA Orientation and Affect Exonuclease Activity.J Am Chem Soc. 2023 Feb 15;145(6):3478-3490. doi: 10.1021/jacs.2c11713. Epub 2023 Feb 6. J Am Chem Soc. 2023. PMID: 36745735 Free PMC article.
-
Dissimilar roles of the four conserved acidic residues in the thermal stability of poly(A)-specific ribonuclease.Int J Mol Sci. 2011;12(5):2901-16. doi: 10.3390/ijms12052901. Epub 2011 May 3. Int J Mol Sci. 2011. PMID: 21686157 Free PMC article. Review.
-
Recent Updates of the CRISPR/Cas9 Genome Editing System: Novel Approaches to Regulate Its Spatiotemporal Control by Genetic and Physicochemical Strategies.Int J Nanomedicine. 2024 Jun 6;19:5335-5363. doi: 10.2147/IJN.S455574. eCollection 2024. Int J Nanomedicine. 2024. PMID: 38859956 Free PMC article. Review.
References
-
- Pelletier H, Sawaya MR, Kumar A, Wilson SH, Kraut J. Structures of ternary complexes of rat DNA polymerase beta, a DNA template-primer, and ddCTP. Science. 1994;264:1891–903. - PubMed
-
- Doublie S, Tabor S, Long AM, Richardson CC, Ellenberger T. Crystal structure of a bacteriophage T7 DNA replication complex at 2.2 A resolution. Nature. 1998;391:251–8. - PubMed
-
- Steitz TA. A mechanism for all polymerases. Nature. 1998;391:231–2. - PubMed
-
- Lee JY, et al. MutH complexed with Hemi- and unmethylated DNAs: coupling base recognition and DNA cleavage. Mol Cell. 2005;20:155–66. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

