The 5' end of two redundant sRNAs is involved in the regulation of multiple targets, including their own regulator
- PMID: 18953042
- PMCID: PMC2588501
- DOI: 10.1093/nar/gkn742
The 5' end of two redundant sRNAs is involved in the regulation of multiple targets, including their own regulator
Abstract
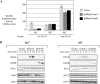
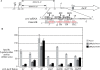
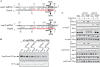
Small RNAs are widespread regulators of gene expression in numerous organisms. This study describes the mode of action of two redundant Escherichia coli sRNAs, OmrA and OmrB, that downregulate the expression of multiple targets, most of which encode outer membrane proteins. Our results show that both sRNAs directly interact with at least two of these target mRNAs, ompT and cirA, in the vicinity of the translation initiation region, consistent with control of these targets being dependent on both Hfq and RNase E. Interestingly, these interactions depend on short stretches of complementarity and involve the conserved 5' end of OmrA/B. A mutation in this region abolishes control of all OmrA/B targets tested thus far, thereby highlighting the crucial role of the OmrA/B 5' end. This allowed us, by looking for mRNA sequences complementary to the OmrA/B 5' end, to identify ompR as an additional direct target of these two sRNAs. Since the OmpR transcriptional regulator activates expression of both omrA and omrB genes, this newly identified control should result in an autoregulatory loop limiting the amount of OmrA/B sRNAs.
Figures




Similar articles
-
Remodelling of the Escherichia coli outer membrane by two small regulatory RNAs.Mol Microbiol. 2006 Jan;59(1):231-47. doi: 10.1111/j.1365-2958.2005.04929.x. Mol Microbiol. 2006. PMID: 16359331
-
Unexpected properties of sRNA promoters allow feedback control via regulation of a two-component system.Nucleic Acids Res. 2016 Nov 16;44(20):9650-9666. doi: 10.1093/nar/gkw642. Epub 2016 Jul 20. Nucleic Acids Res. 2016. PMID: 27439713 Free PMC article.
-
Stem-Loop Structures within mRNA Coding Sequences Activate Translation Initiation and Mediate Control by Small Regulatory RNAs.Mol Cell. 2017 Oct 5;68(1):158-170.e3. doi: 10.1016/j.molcel.2017.08.015. Epub 2017 Sep 14. Mol Cell. 2017. PMID: 28918899
-
Mechanism of RNA silencing by Hfq-binding small RNAs.Curr Opin Microbiol. 2007 Apr;10(2):134-9. doi: 10.1016/j.mib.2007.03.010. Epub 2007 Mar 26. Curr Opin Microbiol. 2007. PMID: 17383928 Review.
-
Small RNAs in the control of RpoS, CsgD, and biofilm architecture of Escherichia coli.RNA Biol. 2014;11(5):494-507. doi: 10.4161/rna.28867. Epub 2014 Apr 25. RNA Biol. 2014. PMID: 25028968 Free PMC article. Review.
Cited by
-
Coherent Feedforward Regulation of Gene Expression by Caulobacter σT and GsrN during Hyperosmotic Stress.J Bacteriol. 2018 Sep 10;200(19):e00349-18. doi: 10.1128/JB.00349-18. Print 2018 Oct 1. J Bacteriol. 2018. PMID: 30012732 Free PMC article.
-
A highly conserved protein of unknown function in Sinorhizobium meliloti affects sRNA regulation similar to Hfq.Nucleic Acids Res. 2011 Jun;39(11):4691-708. doi: 10.1093/nar/gkr060. Epub 2011 Feb 15. Nucleic Acids Res. 2011. PMID: 21325267 Free PMC article.
-
Riboswitch and small RNAs modulate btuB translation initiation in Escherichia coli and trigger distinct mRNA regulatory mechanisms.Nucleic Acids Res. 2024 Jun 10;52(10):5852-5865. doi: 10.1093/nar/gkae347. Nucleic Acids Res. 2024. PMID: 38742638 Free PMC article.
-
Sibling rivalry: related bacterial small RNAs and their redundant and non-redundant roles.Front Cell Infect Microbiol. 2014 Oct 28;4:151. doi: 10.3389/fcimb.2014.00151. eCollection 2014. Front Cell Infect Microbiol. 2014. PMID: 25389522 Free PMC article. Review.
-
Recent Research Advances in Small Regulatory RNAs in Streptococcus.Curr Microbiol. 2021 Jun;78(6):2231-2241. doi: 10.1007/s00284-021-02484-y. Epub 2021 May 7. Curr Microbiol. 2021. PMID: 33963446 Review.
References
-
- Vogel J, Sharma CM. How to find small non-coding RNAs in bacteria. Biol. Chem. 2005;386:1219–1238. - PubMed
-
- Storz G, Gottesman S. In: The RNA World. 3rd. Gesteland RF, Cech TR, Atkins JF, editors. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 2006. pp. 567–594.
-
- Prevost K, Salvail H, Desnoyers G, Jacques JF, Phaneuf E, Massé E. The small RNA RyhB activates the translation of shiA mRNA encoding a permease of shikimate, a compound involved in siderophore synthesis. Mol. Microbiol. 2007;64:1260–1273. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

