Functional targeting of DNA damage to a nuclear pore-associated SUMO-dependent ubiquitin ligase
- PMID: 18948542
- PMCID: PMC3518492
- DOI: 10.1126/science.1162790
Functional targeting of DNA damage to a nuclear pore-associated SUMO-dependent ubiquitin ligase
Abstract
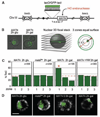
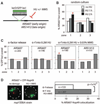
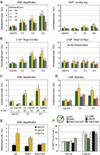
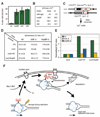
Recent findings suggest important roles for nuclear organization in gene expression. In contrast, little is known about how nuclear organization contributes to genome stability. Epistasis analysis (E-MAP) using DNA repair factors in yeast indicated a functional relationship between a nuclear pore subcomplex and Slx5/Slx8, a small ubiquitin-like modifier (SUMO)-dependent ubiquitin ligase, which we show physically interact. Real-time imaging and chromatin immunoprecipitation confirmed stable recruitment of damaged DNA to nuclear pores. Relocation required the Nup84 complex and Mec1/Tel1 kinases. Spontaneous gene conversion can be enhanced in a Slx8- and Nup84-dependent manner by tethering donor sites at the nuclear periphery. This suggests that strand breaks are shunted to nuclear pores for a repair pathway controlled by a conserved SUMO-dependent E3 ligase.
Figures

Similar articles
-
PolySUMOylation by Siz2 and Mms21 triggers relocation of DNA breaks to nuclear pores through the Slx5/Slx8 STUbL.Genes Dev. 2016 Apr 15;30(8):931-45. doi: 10.1101/gad.277665.116. Epub 2016 Apr 7. Genes Dev. 2016. PMID: 27056668 Free PMC article.
-
Disruption of SUMO-targeted ubiquitin ligases Slx5-Slx8/RNF4 alters RecQ-like helicase Sgs1/BLM localization in yeast and human cells.DNA Repair (Amst). 2015 Feb;26:1-14. doi: 10.1016/j.dnarep.2014.12.004. Epub 2014 Dec 26. DNA Repair (Amst). 2015. PMID: 25588990 Free PMC article.
-
Regulation of recombination at yeast nuclear pores controls repair and triplet repeat stability.Genes Dev. 2015 May 15;29(10):1006-17. doi: 10.1101/gad.256404.114. Epub 2015 May 4. Genes Dev. 2015. PMID: 25940904 Free PMC article.
-
Nuclear organization in genome stability: SUMO connections.Cell Res. 2011 Mar;21(3):474-85. doi: 10.1038/cr.2011.31. Epub 2011 Feb 15. Cell Res. 2011. PMID: 21321608 Free PMC article. Review.
-
SUMO: ligases, isopeptidases and nuclear pores.Trends Biochem Sci. 2003 Nov;28(11):612-8. doi: 10.1016/j.tibs.2003.09.002. Trends Biochem Sci. 2003. PMID: 14607092 Review.
Cited by
-
Rap1 relocalization contributes to the chromatin-mediated gene expression profile and pace of cell senescence.Genes Dev. 2013 Jun 15;27(12):1406-20. doi: 10.1101/gad.218776.113. Epub 2013 Jun 11. Genes Dev. 2013. PMID: 23756653 Free PMC article.
-
Profiling DNA damage-induced phosphorylation in budding yeast reveals diverse signaling networks.Proc Natl Acad Sci U S A. 2016 Jun 28;113(26):E3667-75. doi: 10.1073/pnas.1602827113. Epub 2016 Jun 13. Proc Natl Acad Sci U S A. 2016. PMID: 27298372 Free PMC article.
-
Synthetic protein interactions reveal a functional map of the cell.Elife. 2016 Apr 21;5:e13053. doi: 10.7554/eLife.13053. Elife. 2016. PMID: 27098839 Free PMC article.
-
The Secret Life of Chromosome Loops upon DNA Double-Strand Break.J Mol Biol. 2020 Feb 7;432(3):724-736. doi: 10.1016/j.jmb.2019.07.036. Epub 2019 Aug 8. J Mol Biol. 2020. PMID: 31401119 Free PMC article. Review.
-
Cdk phosphorylation of a nucleoporin controls localization of active genes through the cell cycle.Mol Biol Cell. 2010 Oct 1;21(19):3421-32. doi: 10.1091/mbc.E10-01-0065. Epub 2010 Aug 11. Mol Biol Cell. 2010. PMID: 20702586 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

