Novel control of S phase of the cell cycle by ubiquitin-conjugating enzyme H7
- PMID: 18946090
- PMCID: PMC2613108
- DOI: 10.1091/mbc.e08-01-0036
Novel control of S phase of the cell cycle by ubiquitin-conjugating enzyme H7
Abstract
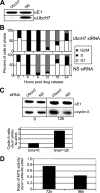
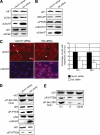
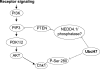
Timely degradation of regulatory proteins by the ubiquitin proteolytic pathway (UPP) is an established paradigm of cell cycle regulation during the G2/M and G1/S transitions. Less is known about roles for the UPP during S phase. Here we present evidence that dynamic cell cycle-dependent changes in levels of UbcH7 regulate entrance into and progression through S phase. In diverse cell lines, UbcH7 protein levels are dramatically reduced in S phase but are fully restored by G2. Knockdown of UbcH7 increases the proportion of cells in S phase and doubles the time to traverse S phase, whereas UbcH7 overexpression reduces the proportion of cells in S phase. These data suggest a role for UbcH7 targets in the completion of S phase and entry into G2. Notably, UbcH7 knockdown was coincident with elevated levels of the checkpoint kinase Chk1 but not Chk2. These results argue that UbcH7 promotes S phase progression to G2 by modulating the intra-S phase checkpoint mediated by Chk1. Furthermore, UbcH7 levels appear to be regulated by a UPP. Together the data identify novel roles for the UPP, specifically UbcH7 in the regulation of S phase transit time as well as in cell proliferation.
Figures



Similar articles
-
Stabilization of p27Kip1/CDKN1B by UBCH7/UBE2L3 catalyzed ubiquitinylation: a new paradigm in cell-cycle control.FASEB J. 2019 Jan;33(1):1235-1247. doi: 10.1096/fj.201800960R. Epub 2018 Aug 16. FASEB J. 2019. PMID: 30113882 Free PMC article.
-
Ubiquitin control of S phase: a new role for the ubiquitin conjugating enzyme, UbcH7.Cell Div. 2009 Aug 7;4:17. doi: 10.1186/1747-1028-4-17. Cell Div. 2009. PMID: 19664228 Free PMC article.
-
Regulation of the ubiquitin proteasome pathway in human lens epithelial cells during the cell cycle.Exp Eye Res. 2004 Feb;78(2):197-205. doi: 10.1016/j.exer.2003.11.009. Exp Eye Res. 2004. PMID: 14729352
-
Chk1 in the DNA damage response: conserved roles from yeasts to mammals.DNA Repair (Amst). 2004 Aug-Sep;3(8-9):1025-32. doi: 10.1016/j.dnarep.2004.03.003. DNA Repair (Amst). 2004. PMID: 15279789 Review.
-
Checking on DNA damage in S phase.Nat Rev Mol Cell Biol. 2004 Oct;5(10):792-804. doi: 10.1038/nrm1493. Nat Rev Mol Cell Biol. 2004. PMID: 15459660 Review.
Cited by
-
Stabilization of p27Kip1/CDKN1B by UBCH7/UBE2L3 catalyzed ubiquitinylation: a new paradigm in cell-cycle control.FASEB J. 2019 Jan;33(1):1235-1247. doi: 10.1096/fj.201800960R. Epub 2018 Aug 16. FASEB J. 2019. PMID: 30113882 Free PMC article.
-
Mechanism and Disease Association With a Ubiquitin Conjugating E2 Enzyme: UBE2L3.Front Immunol. 2022 Feb 21;13:793610. doi: 10.3389/fimmu.2022.793610. eCollection 2022. Front Immunol. 2022. PMID: 35265070 Free PMC article. Review.
-
The efficacy and safety of Collagen-I and hypoxic conditions in urine-derived stem cell ex vivo culture.Tissue Eng Regen Med. 2016 Aug 5;13(4):403-415. doi: 10.1007/s13770-016-9073-6. eCollection 2016 Aug. Tissue Eng Regen Med. 2016. PMID: 30603422 Free PMC article.
-
Mass spectrometry-based quantification of the cellular response to methyl methanesulfonate treatment in human cells.DNA Repair (Amst). 2014 Mar;15:29-38. doi: 10.1016/j.dnarep.2013.12.007. Epub 2014 Jan 22. DNA Repair (Amst). 2014. PMID: 24461736 Free PMC article.
-
Ubiquitin control of S phase: a new role for the ubiquitin conjugating enzyme, UbcH7.Cell Div. 2009 Aug 7;4:17. doi: 10.1186/1747-1028-4-17. Cell Div. 2009. PMID: 19664228 Free PMC article.
References
-
- Anan T., et al. Human ubiquitin-protein ligase Nedd4, expression, subcellular localization and selective interaction with ubiquitin-conjugating enzymes. Genes Cells. 1998;3:751–763. - PubMed
-
- Bornstein G., Bloom J., Sitry-Shevah D., Nakayama K., Pagano M., Hershko A. Role of the SCFSkp2 ubiquitin ligase in the degradation of p21Cip1 in S phase. J. Biol. Chem. 2003;278:25752–25757. - PubMed
-
- Carrano A. C., Eytan E., Hershko A., Pagano M. SKP2 is required for ubiquitin-mediated degradation of the CDK inhibitor p27. Nat. Cell Biol. 1999;1:193–199. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

