Identifying specific protein interaction partners using quantitative mass spectrometry and bead proteomes
- PMID: 18936248
- PMCID: PMC2568020
- DOI: 10.1083/jcb.200805092
Identifying specific protein interaction partners using quantitative mass spectrometry and bead proteomes
Abstract
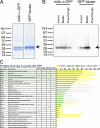
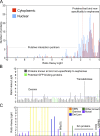
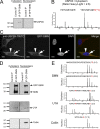
The identification of interaction partners in protein complexes is a major goal in cell biology. Here we present a reliable affinity purification strategy to identify specific interactors that combines quantitative SILAC-based mass spectrometry with characterization of common contaminants binding to affinity matrices (bead proteomes). This strategy can be applied to affinity purification of either tagged fusion protein complexes or endogenous protein complexes, illustrated here using the well-characterized SMN complex as a model. GFP is used as the tag of choice because it shows minimal nonspecific binding to mammalian cell proteins, can be quantitatively depleted from cell extracts, and allows the integration of biochemical protein interaction data with in vivo measurements using fluorescence microscopy. Proteins binding nonspecifically to the most commonly used affinity matrices were determined using quantitative mass spectrometry, revealing important differences that affect experimental design. These data provide a specificity filter to distinguish specific protein binding partners in both quantitative and nonquantitative pull-down and immunoprecipitation experiments.
Figures



Similar articles
-
Establishment of a protein frequency library and its application in the reliable identification of specific protein interaction partners.Mol Cell Proteomics. 2010 May;9(5):861-79. doi: 10.1074/mcp.M900517-MCP200. Epub 2009 Dec 20. Mol Cell Proteomics. 2010. PMID: 20023298 Free PMC article.
-
Protein-Protein Interaction: Tandem Affinity Purification in Bacteria.Methods Mol Biol. 2017;1615:221-232. doi: 10.1007/978-1-4939-7033-9_18. Methods Mol Biol. 2017. PMID: 28667616
-
Intelligent Mixing of Proteomes for Elimination of False Positives in Affinity Purification-Mass Spectrometry.J Proteome Res. 2016 Oct 7;15(10):3929-3937. doi: 10.1021/acs.jproteome.6b00517. Epub 2016 Sep 28. J Proteome Res. 2016. PMID: 27640904
-
Assignment of protein interactions from affinity purification/mass spectrometry data.J Proteome Res. 2012 Mar 2;11(3):1462-74. doi: 10.1021/pr2011632. Epub 2012 Feb 10. J Proteome Res. 2012. PMID: 22283744 Review.
-
Profiling of protein interaction networks of protein complexes using affinity purification and quantitative mass spectrometry.Mol Cell Proteomics. 2010 Aug;9(8):1650-65. doi: 10.1074/mcp.R110.000265. Epub 2010 May 5. Mol Cell Proteomics. 2010. PMID: 20445003 Free PMC article. Review.
Cited by
-
The cellular interactome of the coronavirus infectious bronchitis virus nucleocapsid protein and functional implications for virus biology.J Virol. 2013 Sep;87(17):9486-500. doi: 10.1128/JVI.00321-13. Epub 2013 May 1. J Virol. 2013. PMID: 23637410 Free PMC article.
-
14-3-3ε mediates the cell fate decision-making pathways in response of hepatocellular carcinoma to Bleomycin-induced DNA damage.PLoS One. 2013;8(3):e55268. doi: 10.1371/journal.pone.0055268. Epub 2013 Mar 5. PLoS One. 2013. PMID: 23472066 Free PMC article.
-
Using proteomics to identify host cell interaction partners for VgrG and IglJ.Sci Rep. 2020 Sep 3;10(1):14612. doi: 10.1038/s41598-020-71641-3. Sci Rep. 2020. PMID: 32884055 Free PMC article.
-
The chromatin-associated lncREST ensures effective replication stress response by promoting the assembly of fork signaling factors.Nat Commun. 2024 Feb 1;15(1):978. doi: 10.1038/s41467-024-45183-5. Nat Commun. 2024. PMID: 38302450 Free PMC article.
-
DARPins recognizing mTFP1 as novel reagents for in vitro and in vivo protein manipulations.Biol Open. 2018 Oct 29;7(11):bio036749. doi: 10.1242/bio.036749. Biol Open. 2018. PMID: 30237292 Free PMC article.
References
-
- Al-Hakim, A.K., A. Zagorska, L. Chapman, M. Deak, M. Peggie, and D.R. Alessi. 2008. Control of AMPK-related kinases by USP9X and atypical Lys(29)/Lys(33)-linked polyubiquitin chains. Biochem. J. 411:249–260. - PubMed
-
- Baccon, J., L. Pellizzoni, J. Rappsilber, M. Mann, and G. Dreyfuss. 2002. Identification and characterization of Gemin7, a novel component of the survival of motor neuron complex. J. Biol. Chem. 277:31957–31962. - PubMed
-
- Blagoev, B., I. Kratchmarova, S.E. Ong, M. Nielsen, L.J. Foster, and M. Mann. 2003. A proteomics strategy to elucidate functional protein-protein interactions applied to EGF signaling. Nat. Biotechnol. 21:315–318. - PubMed
-
- Campbell, L., K.M. Hunter, P. Mohaghegh, J.M. Tinsley, M.A. Brasch, and K.E. Davies. 2000. Direct interaction of Smn with dp103, a putative RNA helicase: a role for Smn in transcription regulation? Hum. Mol. Genet. 9:1093–1100. - PubMed
-
- Carissimi, C., J. Baccon, M. Straccia, P. Chiarella, A. Maiolica, A. Sawyer, J. Rappsilber, and L. Pellizzoni. 2005. Unrip is a component of SMN complexes active in snRNP assembly. FEBS Lett. 579:2348–2354. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- 073980/Z/03/Z/WT_/Wellcome Trust/United Kingdom
- 073980/WT_/Wellcome Trust/United Kingdom
- MRC_/Medical Research Council/United Kingdom
- BB/C511613/1/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
- BB/C511572/1/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
LinkOut - more resources
Full Text Sources
Other Literature Sources

