ANGLOR: a composite machine-learning algorithm for protein backbone torsion angle prediction
- PMID: 18923703
- PMCID: PMC2559866
- DOI: 10.1371/journal.pone.0003400
ANGLOR: a composite machine-learning algorithm for protein backbone torsion angle prediction
Abstract
We developed a composite machine-learning based algorithm, called ANGLOR, to predict real-value protein backbone torsion angles from amino acid sequences. The input features of ANGLOR include sequence profiles, predicted secondary structure and solvent accessibility. In a large-scale benchmarking test, the mean absolute error (MAE) of the phi/psi prediction is 28 degrees/46 degrees , which is approximately 10% lower than that generated by software in literature. The prediction is statistically different from a random predictor (or a purely secondary-structure-based predictor) with p-value <1.0 x 10(-300) (or <1.0 x 10(-148)) by Wilcoxon signed rank test. For some residues (ILE, LEU, PRO and VAL) and especially the residues in helix and buried regions, the MAE of phi angles is much smaller (10-20 degrees ) than that in other environments. Thus, although the average accuracy of the ANGLOR prediction is still low, the portion of the accurately predicted dihedral angles may be useful in assisting protein fold recognition and ab initio 3D structure modeling.
Conflict of interest statement
Figures
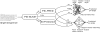
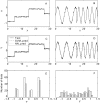

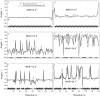
Similar articles
-
TANGLE: two-level support vector regression approach for protein backbone torsion angle prediction from primary sequences.PLoS One. 2012;7(2):e30361. doi: 10.1371/journal.pone.0030361. Epub 2012 Feb 2. PLoS One. 2012. PMID: 22319565 Free PMC article.
-
Real-value prediction of backbone torsion angles.Proteins. 2008 Jul;72(1):427-33. doi: 10.1002/prot.21940. Proteins. 2008. PMID: 18214956
-
Deep learning methods for protein torsion angle prediction.BMC Bioinformatics. 2017 Sep 18;18(1):417. doi: 10.1186/s12859-017-1834-2. BMC Bioinformatics. 2017. PMID: 28923002 Free PMC article.
-
Improving prediction of secondary structure, local backbone angles, and solvent accessible surface area of proteins by iterative deep learning.Sci Rep. 2015 Jun 22;5:11476. doi: 10.1038/srep11476. Sci Rep. 2015. PMID: 26098304 Free PMC article.
-
Sequence based residue depth prediction using evolutionary information and predicted secondary structure.BMC Bioinformatics. 2008 Sep 20;9:388. doi: 10.1186/1471-2105-9-388. BMC Bioinformatics. 2008. PMID: 18803867 Free PMC article.
Cited by
-
The Identification of Metal Ion Ligand-Binding Residues by Adding the Reclassified Relative Solvent Accessibility.Front Genet. 2020 Mar 19;11:214. doi: 10.3389/fgene.2020.00214. eCollection 2020. Front Genet. 2020. PMID: 32265982 Free PMC article.
-
Predicting Ca2+ and Mg2+ ligand binding sites by deep neural network algorithm.BMC Bioinformatics. 2022 Jan 20;22(Suppl 12):324. doi: 10.1186/s12859-021-04250-0. BMC Bioinformatics. 2022. PMID: 35045825 Free PMC article.
-
STRUM: structure-based prediction of protein stability changes upon single-point mutation.Bioinformatics. 2016 Oct 1;32(19):2936-46. doi: 10.1093/bioinformatics/btw361. Epub 2016 Jun 17. Bioinformatics. 2016. PMID: 27318206 Free PMC article.
-
Application of the MAHDS Method for Multiple Alignment of Highly Diverged Amino Acid Sequences.Int J Mol Sci. 2022 Mar 29;23(7):3764. doi: 10.3390/ijms23073764. Int J Mol Sci. 2022. PMID: 35409125 Free PMC article.
-
DeepMSA: constructing deep multiple sequence alignment to improve contact prediction and fold-recognition for distant-homology proteins.Bioinformatics. 2020 Apr 1;36(7):2105-2112. doi: 10.1093/bioinformatics/btz863. Bioinformatics. 2020. PMID: 31738385 Free PMC article.
References
-
- Branden C, Tooze J. 1999. Introduction to protein structure: Garland Publishing, Inc.
-
- Neal S, Berjanskii M, Zhang H, Wishart DS. Accurate prediction of protein torsion angles using chemical shifts and sequence homology. Magn Reson Chem 44 Spec No. 2006:S158–167. - PubMed
-
- Wood MJ, Hirst JD. Protein secondary structure prediction with dihedral angles. Proteins. 2005;59:476–481. - PubMed
-
- Mooney C, Vullo A, Pollastri G. Protein structural motif prediction in multidimensional phi-psi space leads to improved secondary structure prediction. J Comput Biol. 2006;13:1489–1502. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

