T cell responses to whole SARS coronavirus in humans
- PMID: 18832706
- PMCID: PMC2683413
- DOI: 10.4049/jimmunol.181.8.5490
T cell responses to whole SARS coronavirus in humans
Abstract
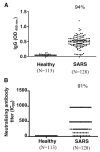
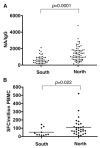
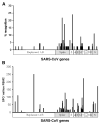
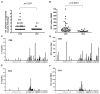
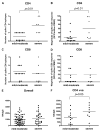
Effective vaccines should confer long-term protection against future outbreaks of severe acute respiratory syndrome (SARS) caused by a novel zoonotic coronavirus (SARS-CoV) with unknown animal reservoirs. We conducted a cohort study examining multiple parameters of immune responses to SARS-CoV infection, aiming to identify the immune correlates of protection. We used a matrix of overlapping peptides spanning whole SARS-CoV proteome to determine T cell responses from 128 SARS convalescent samples by ex vivo IFN-gamma ELISPOT assays. Approximately 50% of convalescent SARS patients were positive for T cell responses, and 90% possessed strongly neutralizing Abs. Fifty-five novel T cell epitopes were identified, with spike protein dominating total T cell responses. CD8(+) T cell responses were more frequent and of a greater magnitude than CD4(+) T cell responses (p < 0.001). Polychromatic cytometry analysis indicated that the virus-specific T cells from the severe group tended to be a central memory phenotype (CD27(+)/CD45RO(+)) with a significantly higher frequency of polyfunctional CD4(+) T cells producing IFN-gamma, TNF-alpha, and IL-2, and CD8(+) T cells producing IFN-gamma, TNF-alpha, and CD107a (degranulation), as compared with the mild-moderate group. Strong T cell responses correlated significantly (p < 0.05) with higher neutralizing Ab. The serum cytokine profile during acute infection indicated a significant elevation of innate immune responses. Increased Th2 cytokines were observed in patients with fatal infection. Our study provides a roadmap for the immunogenicity of SARS-CoV and types of immune responses that may be responsible for the virus clearance, and should serve as a benchmark for SARS-CoV vaccine design and evaluation.
Conflict of interest statement
The authors have no financial conflict of interest.
Figures



Similar articles
-
Virus-specific memory CD8 T cells provide substantial protection from lethal severe acute respiratory syndrome coronavirus infection.J Virol. 2014 Oct;88(19):11034-44. doi: 10.1128/JVI.01505-14. Epub 2014 Jul 23. J Virol. 2014. PMID: 25056892 Free PMC article.
-
CD8+ T cell response in HLA-A*0201 transgenic mice is elicited by epitopes from SARS-CoV S protein.Vaccine. 2010 Sep 24;28(41):6666-74. doi: 10.1016/j.vaccine.2010.08.013. Epub 2010 Aug 13. Vaccine. 2010. PMID: 20709007 Free PMC article.
-
Priming with SARS CoV S DNA and boosting with SARS CoV S epitopes specific for CD4+ and CD8+ T cells promote cellular immune responses.Vaccine. 2007 Sep 28;25(39-40):6981-91. doi: 10.1016/j.vaccine.2007.06.047. Epub 2007 Jul 16. Vaccine. 2007. PMID: 17709158 Free PMC article.
-
SARS Immunity and Vaccination.Cell Mol Immunol. 2004 Jun;1(3):193-8. Cell Mol Immunol. 2004. PMID: 16219167 Review.
-
T cell-mediated immune response to respiratory coronaviruses.Immunol Res. 2014 Aug;59(1-3):118-28. doi: 10.1007/s12026-014-8534-z. Immunol Res. 2014. PMID: 24845462 Free PMC article. Review.
Cited by
-
Evolution of SARS-CoV-2: Review of Mutations, Role of the Host Immune System.Nephron. 2021;145(4):392-403. doi: 10.1159/000515417. Epub 2021 Apr 28. Nephron. 2021. PMID: 33910211 Free PMC article. Review.
-
Preceding infection and risk of stroke: An old concept revived by the COVID-19 pandemic.Int J Stroke. 2020 Oct;15(7):722-732. doi: 10.1177/1747493020943815. Epub 2020 Jul 24. Int J Stroke. 2020. PMID: 32618498 Free PMC article. Review.
-
Neurological Complications Associated with the Blood-Brain Barrier Damage Induced by the Inflammatory Response During SARS-CoV-2 Infection.Mol Neurobiol. 2021 Feb;58(2):520-535. doi: 10.1007/s12035-020-02134-7. Epub 2020 Sep 25. Mol Neurobiol. 2021. PMID: 32978729 Free PMC article. Review.
-
Th2 and Th17-associated immunopathology following SARS-CoV-2 breakthrough infection in Spike-vaccinated ACE2-humanized mice.J Med Virol. 2024 Jan;96(1):e29408. doi: 10.1002/jmv.29408. J Med Virol. 2024. PMID: 38258331 Free PMC article.
-
The Long Road Toward COVID-19 Herd Immunity: Vaccine Platform Technologies and Mass Immunization Strategies.Front Immunol. 2020 Jul 21;11:1817. doi: 10.3389/fimmu.2020.01817. eCollection 2020. Front Immunol. 2020. PMID: 32793245 Free PMC article. Review.
References
-
- Li W, Shi Z, Yu M, Ren W, Smith C, Epstein JH, Wang H, Crameri G, Hu Z, Zhang H, et al. Bats are natural reservoirs of SARS-like corona-viruses. Science. 2005;310:676–679. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

