Chromosome-scale genetic mapping using a set of 16 conditionally stable Saccharomyces cerevisiae chromosomes
- PMID: 18832360
- PMCID: PMC2600922
- DOI: 10.1534/genetics.108.087999
Chromosome-scale genetic mapping using a set of 16 conditionally stable Saccharomyces cerevisiae chromosomes
Abstract
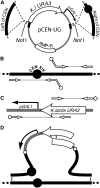
We have created a resource to rapidly map genetic traits to specific chromosomes in yeast. This mapping is done using a set of 16 yeast strains each containing a different chromosome with a conditionally functional centromere. Conditional centromere function is achieved by integration of a GAL1 promoter in cis to centromere sequences. We show that the 16 yeast chromosomes can be individually lost in diploid strains, which become hemizygous for the destabilized chromosome. Interestingly, most 2n - 1 strains endoduplicate and become 2n. We also demonstrate how chromosome loss in this set of strains can be used to map both recessive and dominant markers to specific chromosomes. In addition, we show that this method can be used to rapidly validate gene assignments from screens of strain libraries such as the yeast gene disruption collection.
Figures



Similar articles
-
Chromosome V loss due to centromere knockout or MAD2-deletion is immediately followed by restitution of homozygous diploidy in Saccharomyces cerevisiae.Yeast. 2002 Apr;19(6):553-64. doi: 10.1002/yea.859. Yeast. 2002. PMID: 11921104
-
Chromosomal assignment of mutations by specific chromosome loss in the yeast Saccharomyces cerevisiae.Genetics. 1990 Jun;125(2):333-40. doi: 10.1093/genetics/125.2.333. Genetics. 1990. PMID: 2199315 Free PMC article.
-
Systematic hybrid LOH: a new method to reduce false positives and negatives during screening of yeast gene deletion libraries.Yeast. 2006 Oct-Nov;23(14-15):1097-106. doi: 10.1002/yea.1423. Yeast. 2006. PMID: 17083134
-
Exploring genetic interactions and networks with yeast.Nat Rev Genet. 2007 Jun;8(6):437-49. doi: 10.1038/nrg2085. Nat Rev Genet. 2007. PMID: 17510664 Review.
-
The centromere of budding yeast.Bioessays. 1993 Jul;15(7):451-60. doi: 10.1002/bies.950150704. Bioessays. 1993. PMID: 8379948 Review.
Cited by
-
Rapid and Efficient CRISPR/Cas9-Based Mating-Type Switching of Saccharomyces cerevisiae.G3 (Bethesda). 2018 Jan 4;8(1):173-183. doi: 10.1534/g3.117.300347. G3 (Bethesda). 2018. PMID: 29150593 Free PMC article.
-
Context-dependent neocentromere activity in synthetic yeast chromosome VIII.Cell Genom. 2023 Nov 9;3(11):100437. doi: 10.1016/j.xgen.2023.100437. eCollection 2023 Nov 8. Cell Genom. 2023. PMID: 38020969 Free PMC article.
-
Actin dosage lethality screening in yeast mediated by selective ploidy ablation reveals links to urmylation/wobble codon recognition and chromosome stability.G3 (Bethesda). 2013 Mar;3(3):553-61. doi: 10.1534/g3.113.005579. Epub 2013 Mar 1. G3 (Bethesda). 2013. PMID: 23450344 Free PMC article.
-
Most, but not all, yeast strains in the deletion library contain the [PIN(+)] prion.Yeast. 2010 Mar;27(3):159-66. doi: 10.1002/yea.1740. Yeast. 2010. PMID: 20014044 Free PMC article.
-
A Cohesin-Based Partitioning Mechanism Revealed upon Transcriptional Inactivation of Centromere.PLoS Genet. 2016 Apr 29;12(4):e1006021. doi: 10.1371/journal.pgen.1006021. eCollection 2016 Apr. PLoS Genet. 2016. PMID: 27128635 Free PMC article.
References
-
- Chlebowicz-Sledziewska, E., and A. Z. Sledziewski, 1985. Construction of multicopy yeast plasmids with regulated centromere function. Gene 39 25–31. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- GM39067/GM/NIGMS NIH HHS/United States
- R33 CA125520-02/CA/NCI NIH HHS/United States
- R01 GM039067/GM/NIGMS NIH HHS/United States
- GM57254/GM/NIGMS NIH HHS/United States
- HG00193/HG/NHGRI NIH HHS/United States
- R01 HG001620/HG/NHGRI NIH HHS/United States
- R37 GM050237/GM/NIGMS NIH HHS/United States
- R01 GM050237/GM/NIGMS NIH HHS/United States
- CA125520/CA/NCI NIH HHS/United States
- R01 GM048624/GM/NIGMS NIH HHS/United States
- HG01620/HG/NHGRI NIH HHS/United States
- R33 CA125520/CA/NCI NIH HHS/United States
- GM48624/GM/NIGMS NIH HHS/United States
- GM50237/GM/NIGMS NIH HHS/United States
- F32 HG000193/HG/NHGRI NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

