Notch1 regulates the fate of cardiac progenitor cells
- PMID: 18832173
- PMCID: PMC2563067
- DOI: 10.1073/pnas.0808357105
Notch1 regulates the fate of cardiac progenitor cells
Retraction in
-
Retraction for Boni et al., Notch1 regulates the fate of cardiac progenitor cells.Proc Natl Acad Sci U S A. 2019 Oct 8;116(41):20796. doi: 10.1073/pnas.1916178116. Epub 2019 Sep 30. Proc Natl Acad Sci U S A. 2019. PMID: 31570570 Free PMC article. No abstract available.
Abstract
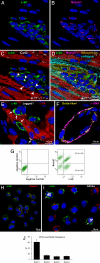
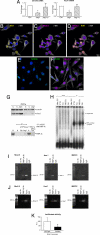
The Notch receptor mediates cell fate decision in multiple organs. In the current work we tested the hypothesis that Nkx2.5 is a target gene of Notch1 and raised the possibility that Notch1 regulates myocyte commitment in the adult heart. Cardiac progenitor cells (CPCs) in the niches express Notch1 receptor, and the supporting cells exhibit the Notch ligand Jagged1. The nuclear translocation of Notch1 intracellular domain (N1ICD) up-regulates Nkx2.5 in CPCs and promotes the formation of cycling myocytes in vitro. N1ICD and RBP-Jk form a protein complex, which in turn binds to the Nkx2.5 promoter initiating transcription and myocyte differentiation. In contrast, transcription factors of vascular cells are down-regulated by Jagged1 activation of the Notch1 pathway. Importantly, inhibition of Notch1 in infarcted mice impairs the commitment of resident CPCs to the myocyte lineage opposing cardiomyogenesis. These observations indicate that Notch1 favors the early specification of CPCs to the myocyte phenotype but maintains the newly formed cells in a highly proliferative state. Dividing Nkx2.5-positive myocytes correspond to transit amplifying cells, which condition the replicative capacity of the heart. In conclusion, Notch1 may have critical implications in the control of heart homeostasis and its adaptation to pathologic states.
Conflict of interest statement
The authors declare no conflict of interest.
Figures



Similar articles
-
Activation of Notch1 signalling promotes multi-lineage differentiation of c-Kit(POS)/NKX2.5(POS) bone marrow stem cells: implication in stem cell translational medicine.Stem Cell Res Ther. 2015 May 9;6(1):91. doi: 10.1186/s13287-015-0085-2. Stem Cell Res Ther. 2015. PMID: 25956503 Free PMC article.
-
Directed differentiation of patient-specific induced pluripotent stem cells identifies the transcriptional repression and epigenetic modification of NKX2-5, HAND1, and NOTCH1 in hypoplastic left heart syndrome.PLoS One. 2014 Jul 22;9(7):e102796. doi: 10.1371/journal.pone.0102796. eCollection 2014. PLoS One. 2014. PMID: 25050861 Free PMC article.
-
Transcription factor-induced activation of cardiac gene expression in human c-kit+ cardiac progenitor cells.PLoS One. 2017 Mar 29;12(3):e0174242. doi: 10.1371/journal.pone.0174242. eCollection 2017. PLoS One. 2017. PMID: 28355297 Free PMC article.
-
Cardiac stem cell niches.Stem Cell Res. 2014 Nov;13(3 Pt B):631-46. doi: 10.1016/j.scr.2014.09.001. Epub 2014 Sep 8. Stem Cell Res. 2014. PMID: 25267073 Free PMC article. Review.
-
Role of stem cells in cardiovascular biology.J Thromb Haemost. 2011 Jul;9 Suppl 1(0 1):151-61. doi: 10.1111/j.1538-7836.2011.04363.x. J Thromb Haemost. 2011. PMID: 21781250 Free PMC article. Review.
Cited by
-
Deciphering Cardiac Biology and Disease by Single-Cell Transcriptomic Profiling.Biomolecules. 2022 Apr 12;12(4):566. doi: 10.3390/biom12040566. Biomolecules. 2022. PMID: 35454155 Free PMC article. Review.
-
Cardiac transcription factors driven lineage-specification of adult stem cells.J Cardiovasc Transl Res. 2010 Feb;3(1):61-5. doi: 10.1007/s12265-009-9144-3. Epub 2009 Oct 21. J Cardiovasc Transl Res. 2010. PMID: 20560034
-
Peptide-Based Functional Biomaterials for Soft-Tissue Repair.Front Bioeng Biotechnol. 2019 Aug 23;7:205. doi: 10.3389/fbioe.2019.00205. eCollection 2019. Front Bioeng Biotechnol. 2019. PMID: 31508416 Free PMC article. Review.
-
Spontaneous calcium oscillations regulate human cardiac progenitor cell growth.Circ Res. 2009 Oct 9;105(8):764-74. doi: 10.1161/CIRCRESAHA.109.206698. Epub 2009 Sep 10. Circ Res. 2009. PMID: 19745162 Free PMC article.
-
Prometheus's heart: what lies beneath.J Cell Mol Med. 2012 Feb;16(2):228-36. doi: 10.1111/j.1582-4934.2011.01487.x. J Cell Mol Med. 2012. PMID: 22099480 Free PMC article.
References
-
- Bray SJ. Notch signaling: A simple pathway becomes complex. Nat Rev Mol Cell Biol. 2006;7:678–689. - PubMed
-
- Tanigaki K, Honjo T. Regulation of lymphocyte development by Notch signaling. Nat Immunol. 2007;8:451–456. - PubMed
-
- Iso T, Kades L, Hamamori Y. HES and HERP families: Multiple effector of the Notch signaling pathway. J Cell Physiol. 2003;194:237–255. - PubMed
-
- Estrach S, Ambler CA, Lo Celso C, Hozumi K, Watt FM. Jagged1 is a beta-catenin target gene required for ectopic hair follicle formation in adult epidermis. Development. 2006;133:4427–4438. - PubMed
-
- Calvi LM, et al. Osteoblastic cells regulate the hematopoietic stem cell niche. Nature. 2003;425:841–846. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

