Epigenetic silencing of human immunodeficiency virus (HIV) transcription by formation of restrictive chromatin structures at the viral long terminal repeat drives the progressive entry of HIV into latency
- PMID: 18829756
- PMCID: PMC2593349
- DOI: 10.1128/JVI.01383-08
Epigenetic silencing of human immunodeficiency virus (HIV) transcription by formation of restrictive chromatin structures at the viral long terminal repeat drives the progressive entry of HIV into latency
Abstract
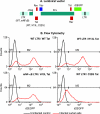
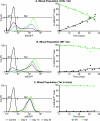
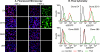
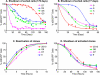
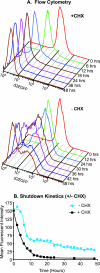
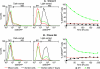
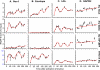
The molecular mechanisms utilized by human immunodeficiency virus (HIV) to enter latency are poorly understood. Following the infection of Jurkat T cells with lentiviral vectors that express Tat in cis, gene expression is progressively silenced. Silencing is greatly enhanced when the lentiviral vectors carry an attenuated Tat gene with the H13L mutation. Individual clones of lentivirus-infected cells showed a wide range of shutdown rates, with the majority showing a 50% silencing frequency between 30 to 80 days. The silenced clones characteristically contained a small fraction (0 to 15%) of activated cells that continued to express d2EGFP. When d2EGFP(+) and d2EGFP(-) cell populations were isolated from the shutdown clones, they quickly reverted to the original distribution of inactive and active cells, suggesting that the d2EGFP(+) cells arise from stochastic fluctuations in gene expression. The detailed analysis of transcription initiation and elongation using chromatin immunoprecipitation (ChIP) assays confirms that Tat levels are restricted in the latently infected cells but gradually rise during proviral reactivation. ChIP assays using clones of latently infected cells demonstrate that the latent proviruses carry high levels of deacetylated histones and trimethylated histones. In contrast, the cellular genes IkappaB alpha and GAPDH had high levels of acetylated histones and no trimethylated histones. The levels of trimethylated histone H3 and HP1-alpha associated with HIV proviruses fell rapidly after tumor necrosis factor alpha activation. The progressive shutdown of HIV transcription following infection suggests that epigenetic mechanisms targeting chromatin structures selectively restrict HIV transcription initiation. This decreases Tat production below the levels that are required to sustain HIV gene expression.
Figures
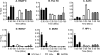
Similar articles
-
Establishment of HIV latency in primary CD4+ cells is due to epigenetic transcriptional silencing and P-TEFb restriction.J Virol. 2010 Jul;84(13):6425-37. doi: 10.1128/JVI.01519-09. Epub 2010 Apr 21. J Virol. 2010. PMID: 20410271 Free PMC article.
-
Sustained induction of NF-kappa B is required for efficient expression of latent human immunodeficiency virus type 1.J Virol. 2007 Jun;81(11):6043-56. doi: 10.1128/JVI.02074-06. Epub 2007 Mar 21. J Virol. 2007. PMID: 17376917 Free PMC article.
-
Chromatin-associated regulation of HIV-1 transcription: implications for the development of therapeutic strategies.Subcell Biochem. 2007;41:371-96. Subcell Biochem. 2007. PMID: 17484137 Review.
-
Shutdown of HIV-1 Transcription in T Cells by Nullbasic, a Mutant Tat Protein.mBio. 2016 Jul 5;7(4):e00518-16. doi: 10.1128/mBio.00518-16. mBio. 2016. PMID: 27381288 Free PMC article.
-
Transcription of HIV: Tat and cellular chromatin.Adv Pharmacol. 2007;55:137-59. doi: 10.1016/S1054-3589(07)55004-0. Adv Pharmacol. 2007. PMID: 17586314 Review. No abstract available.
Cited by
-
A Stronger Transcription Regulatory Circuit of HIV-1C Drives the Rapid Establishment of Latency with Implications for the Direct Involvement of Tat.J Virol. 2020 Sep 15;94(19):e00503-20. doi: 10.1128/JVI.00503-20. Print 2020 Sep 15. J Virol. 2020. PMID: 32669338 Free PMC article.
-
Differences in HIV burden and immune activation within the gut of HIV-positive patients receiving suppressive antiretroviral therapy.J Infect Dis. 2010 Nov 15;202(10):1553-61. doi: 10.1086/656722. Epub 2010 Oct 12. J Infect Dis. 2010. PMID: 20939732 Free PMC article. Clinical Trial.
-
Establishment, Persistence, and Reactivation of Latent HIV-1 Infection in Renal Epithelial Cells.J Virol. 2022 Jul 27;96(14):e0062422. doi: 10.1128/jvi.00624-22. Epub 2022 Jul 5. J Virol. 2022. PMID: 35867560 Free PMC article.
-
Transcriptional Circuit Fragility Influences HIV Proviral Fate.Cell Rep. 2019 Apr 2;27(1):154-171.e9. doi: 10.1016/j.celrep.2019.03.007. Cell Rep. 2019. PMID: 30943398 Free PMC article.
-
Transcriptional Stochasticity as a Key Aspect of HIV-1 Latency.Viruses. 2023 Sep 21;15(9):1969. doi: 10.3390/v15091969. Viruses. 2023. PMID: 37766375 Free PMC article. Review.
References
-
- Brooks, D. G., S. G. Kitchen, C. M. Kitchen, D. D. Scripture-Adams, and J. A. Zack. 2001. Generation of HIV latency during thymopoiesis. Nat. Med. 7459-464. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

