A screen of cell-surface molecules identifies leucine-rich repeat proteins as key mediators of synaptic target selection
- PMID: 18817735
- PMCID: PMC2630283
- DOI: 10.1016/j.neuron.2008.07.037
A screen of cell-surface molecules identifies leucine-rich repeat proteins as key mediators of synaptic target selection
Abstract
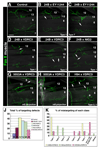
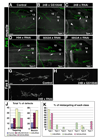
In Drosophila embryos and larvae, a small number of identified motor neurons innervate body wall muscles in a highly stereotyped pattern. Although genetic screens have identified many proteins that are required for axon guidance and synaptogenesis in this system, little is known about the mechanisms by which muscle fibers are defined as targets for specific motor axons. To identify potential target labels, we screened 410 genes encoding cell-surface and secreted proteins, searching for those whose overexpression on all muscle fibers causes motor axons to make targeting errors. Thirty such genes were identified, and a number of these were members of a large gene family encoding proteins whose extracellular domains contain leucine-rich repeat (LRR) sequences, which are protein interaction modules. By manipulating gene expression in muscle 12, we showed that four LRR proteins participate in the selection of this muscle as the appropriate synaptic target for the RP5 motor neuron.
Figures


Similar articles
-
Target recognition and synaptogenesis by motor axons: responses to the sidestep protein.Int J Dev Neurosci. 2005 Jun;23(4):397-410. doi: 10.1016/j.ijdevneu.2004.10.002. Epub 2004 Nov 21. Int J Dev Neurosci. 2005. PMID: 15927764
-
Deconstruction of the beaten Path-Sidestep interaction network provides insights into neuromuscular system development.Elife. 2017 Aug 15;6:e28111. doi: 10.7554/eLife.28111. Elife. 2017. PMID: 28829740 Free PMC article.
-
sidestep encodes a target-derived attractant essential for motor axon guidance in Drosophila.Cell. 2001 Apr 6;105(1):57-67. doi: 10.1016/s0092-8674(01)00296-3. Cell. 2001. PMID: 11301002
-
Motor axon guidance in Drosophila.Semin Cell Dev Biol. 2019 Jan;85:36-47. doi: 10.1016/j.semcdb.2017.11.013. Epub 2017 Nov 24. Semin Cell Dev Biol. 2019. PMID: 29155221 Review.
-
No sidesteps on a beaten track: motor axons follow a labeled substrate pathway.Cell Adh Migr. 2009 Oct-Dec;3(4):358-60. doi: 10.4161/cam.3.4.9491. Epub 2009 Oct 10. Cell Adh Migr. 2009. PMID: 19717972 Free PMC article. Review.
Cited by
-
An extracellular interactome of immunoglobulin and LRR proteins reveals receptor-ligand networks.Cell. 2013 Jul 3;154(1):228-39. doi: 10.1016/j.cell.2013.06.006. Cell. 2013. PMID: 23827685 Free PMC article.
-
Setting up presynaptic structures at specific positions.Curr Opin Neurobiol. 2010 Aug;20(4):489-93. doi: 10.1016/j.conb.2010.04.011. Epub 2010 May 12. Curr Opin Neurobiol. 2010. PMID: 20471244 Free PMC article. Review.
-
Temporal evolution of single-cell transcriptomes of Drosophila olfactory projection neurons.Elife. 2021 Jan 11;10:e63450. doi: 10.7554/eLife.63450. Elife. 2021. PMID: 33427646 Free PMC article.
-
TMEM184b Promotes Axon Degeneration and Neuromuscular Junction Maintenance.J Neurosci. 2016 Apr 27;36(17):4681-9. doi: 10.1523/JNEUROSCI.2893-15.2016. J Neurosci. 2016. PMID: 27122027 Free PMC article.
-
Visualization of binding patterns for five Leucine-rich repeat proteins in the Drosophila embryo.MicroPubl Biol. 2019 Dec 18;2019:10.17912/micropub.biology.000199. doi: 10.17912/micropub.biology.000199. MicroPubl Biol. 2019. PMID: 32550403 Free PMC article. No abstract available.
References
-
- Abrell S, Jackle H. Axon guidance of Drosophila SNb motoneurons depends on the cooperative action of muscular Kruppel and neuronal capricious activities. Mech Dev. 2001;109:3–12. - PubMed
-
- Artero R, Furlong EE, Beckett K, Scott MP, Baylies M. Notch and Ras signaling pathway effector genes expressed in fusion competent and founder cells during Drosophila myogenesis. Development. 2003;130:6257–6272. - PubMed
-
- Brand AH, Perrimon N. Targeted gene expression as a means of altering cell fates and generating dominant phenotypes. Development. 1993;118:401–415. - PubMed
-
- Chang Z, Price BD, Bockheim S, Boedigheimer MJ, Smith R, Laughon A. Molecular and genetic characterization of the Drosophila tartan gene. Dev Biol. 1993;160:315–332. - PubMed
-
- Chiba A, Snow P, Keshishian H, Hotta Y. Fasciclin III as a synaptic target recognition molecule in Drosophila. Nature. 1995;374:166–168. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

