The p110 delta of PI3K plays a critical role in NK cell terminal maturation and cytokine/chemokine generation
- PMID: 18809712
- PMCID: PMC2556795
- DOI: 10.1084/jem.20072327
The p110 delta of PI3K plays a critical role in NK cell terminal maturation and cytokine/chemokine generation
Abstract
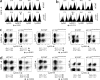
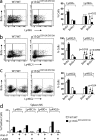
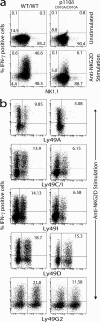
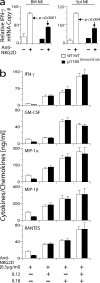
Phosphatidylinositol 3-kinases (PI3Ks) play a critical role in regulating B cell receptor- and T cell receptor-mediated signaling. However, their role in natural killer (NK) cell development and functions is not well understood. Using mice expressing p110 delta(D910A), a catalytically inactive p110 delta, we show that these mice had reduced NK cellularity, defective Ly49C and Ly49I NK subset maturation, and decreased CD27(High) NK numbers. p110 delta inactivation marginally impaired NK-mediated cytotoxicity against tumor cells in vitro and in vivo. However, NKG2D, Ly49D, and NK1.1 receptor-mediated cytokine and chemokine generation by NK cells was severely affected in these mice. Further, p110 delta(D910A/D910A) NK cell-mediated antiviral responses through natural cytotoxicity receptor 1 were reduced. Analysis of signaling events demonstrates that p110 delta(D910A/D910A) NK cells had a reduced c-Jun N-terminal kinase 1/2 phosphorylation in response to NKG2D-mediated activation. These results reveal a previously unrecognized role of PI3K-p110 delta in NK cell development and effector functions.
Figures





Similar articles
-
A requirement for CD45 distinguishes Ly49D-mediated cytokine and chemokine production from killing in primary natural killer cells.J Exp Med. 2005 May 2;201(9):1421-33. doi: 10.1084/jem.20042294. J Exp Med. 2005. PMID: 15867094 Free PMC article.
-
Bcl10 plays a divergent role in NK cell-mediated cytotoxicity and cytokine generation.J Immunol. 2007 Sep 15;179(6):3752-62. doi: 10.4049/jimmunol.179.6.3752. J Immunol. 2007. PMID: 17785812
-
The role of LY49 NK cell subsets in the regulation of murine cytomegalovirus infections.J Immunol. 1999 Jan 15;162(2):718-26. J Immunol. 1999. PMID: 9916691
-
Mouse Ly49 NK receptors: balancing activation and inhibition.Mol Immunol. 2005 Feb;42(4):445-50. doi: 10.1016/j.molimm.2004.07.024. Mol Immunol. 2005. PMID: 15607796 Review.
-
Activation of natural killer cells: underlying molecular mechanisms revealed.Scand J Immunol. 2004 Jul-Aug;60(1-2):14-22. doi: 10.1111/j.0300-9475.2004.01475.x. Scand J Immunol. 2004. PMID: 15238069 Review.
Cited by
-
Functional dichotomy between NKG2D and CD28-mediated co-stimulation in human CD8+ T cells.PLoS One. 2010 Sep 9;5(9):e12635. doi: 10.1371/journal.pone.0012635. PLoS One. 2010. PMID: 20844584 Free PMC article.
-
Disrupted PI3K p110δ Signaling Dysregulates Maternal Immune Cells and Increases Fetal Mortality In Mice.Cell Rep. 2015 Dec 29;13(12):2817-28. doi: 10.1016/j.celrep.2015.11.050. Epub 2015 Dec 17. Cell Rep. 2015. PMID: 26711346 Free PMC article.
-
Role of inositol phospholipid signaling in natural killer cell biology.Front Immunol. 2013 Mar 6;4:47. doi: 10.3389/fimmu.2013.00047. eCollection 2013. Front Immunol. 2013. PMID: 23508471 Free PMC article.
-
IQGAP1: a regulator of intracellular spacetime relativity.J Immunol. 2012 Mar 1;188(5):2057-63. doi: 10.4049/jimmunol.1102439. J Immunol. 2012. PMID: 22345702 Free PMC article. Review.
-
The functional impairment of natural killer cells during influenza virus infection.Immunol Cell Biol. 2009 Nov-Dec;87(8):579-89. doi: 10.1038/icb.2009.60. Epub 2009 Sep 1. Immunol Cell Biol. 2009. PMID: 19721456 Free PMC article.
References
-
- Lanier, L.L. 2005. NK cell recognition. Annu. Rev. Immunol. 23:225–274. - PubMed
-
- Cosman, D., J. Mullberg, C.L. Sutherland, W. Chin, R. Armitage, W. Fanslow, M. Kubin, and N.J. Chalupny. 2001. ULBPs, novel MHC class I-related molecules, bind to CMV glycoprotein UL16 and stimulate NK cytotoxicity through the NKG2D receptor. Immunity. 14:123–133. - PubMed
-
- Diefenbach, A., A.M. Jamieson, S.D. Liu, N. Shastri, and D.H. Raulet. 2000. Ligands for the murine NKG2D receptor: expression by tumor cells and activation of NK cells and macrophages. Nat. Immunol. 1:119–126. - PubMed
-
- Cerwenka, A., A.B. Bakker, T. McClanahan, J. Wagner, J. Wu, J.H. Phillips, and L.L. Lanier. 2000. Retinoic acid early inducible genes define a ligand family for the activating NKG2D receptor in mice. Immunity. 12:721–727. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

