Effect of proliferating cell nuclear antigen ubiquitination and chromatin structure on the dynamic properties of the Y-family DNA polymerases
- PMID: 18799611
- PMCID: PMC2592664
- DOI: 10.1091/mbc.e08-07-0724
Effect of proliferating cell nuclear antigen ubiquitination and chromatin structure on the dynamic properties of the Y-family DNA polymerases
Abstract
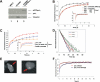
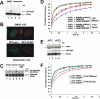
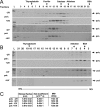
Y-family DNA polymerases carry out translesion synthesis past damaged DNA. DNA polymerases (pol) eta and iota are usually uniformly distributed through the nucleus but accumulate in replication foci during S phase. DNA-damaging treatments result in an increase in S phase cells containing polymerase foci. Using photobleaching techniques, we show that poleta is highly mobile in human fibroblasts. Even when localized in replication foci, it is only transiently immobilized. Although ubiquitination of proliferating cell nuclear antigen (PCNA) is not required for the localization of poleta in foci, it results in an increased residence time in foci. poliota is even more mobile than poleta, both when uniformly distributed and when localized in foci. Kinetic modeling suggests that both poleta and poliota diffuse through the cell but that they are transiently immobilized for approximately 150 ms, with a larger proportion of poleta than poliota immobilized at any time. Treatment of cells with DRAQ5, which results in temporary opening of the chromatin structure, causes a dramatic immobilization of poleta but not poliota. Our data are consistent with a model in which the polymerases are transiently probing the DNA/chromatin. When DNA is exposed at replication forks, the polymerase residence times increase, and this is further facilitated by the ubiquitination of PCNA.
Figures
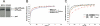


Similar articles
-
Localization of DNA polymerases eta and iota to the replication machinery is tightly co-ordinated in human cells.EMBO J. 2003 Mar 3;22(5):1223-33. doi: 10.1093/emboj/cdf618. EMBO J. 2003. PMID: 12606586 Free PMC article.
-
Proliferating cell nuclear antigen-dependent coordination of the biological functions of human DNA polymerase iota.J Biol Chem. 2004 Nov 12;279(46):48360-8. doi: 10.1074/jbc.M406511200. Epub 2004 Sep 1. J Biol Chem. 2004. PMID: 15342632
-
Localization of DNA polymerases eta and iota to the replication machinery is tightly co-ordinated in human cells.EMBO J. 2002 Nov 15;21(22):6246-56. doi: 10.1093/emboj/cdf618. EMBO J. 2002. Corrected and republished in: EMBO J. 2003 Mar 3;22(5):1223-33. doi: 10.1093/emboj/cdf618. PMID: 12426396 Free PMC article. Corrected and republished.
-
poliota-dependent lesion bypass in vitro.Mutat Res. 2002 Dec 29;510(1-2):9-22. doi: 10.1016/s0027-5107(02)00248-8. Mutat Res. 2002. PMID: 12459439 Review.
-
Ubiquitin-dependent regulation of translesion polymerases.Biochem Soc Trans. 2010 Feb;38(Pt 1):110-5. doi: 10.1042/BST0380110. Biochem Soc Trans. 2010. PMID: 20074045 Review.
Cited by
-
FF483-484 motif of human Polη mediates its interaction with the POLD2 subunit of Polδ and contributes to DNA damage tolerance.Nucleic Acids Res. 2015 Feb 27;43(4):2116-25. doi: 10.1093/nar/gkv076. Epub 2015 Feb 6. Nucleic Acids Res. 2015. PMID: 25662213 Free PMC article.
-
Regulation of PCNA-protein interactions for genome stability.Nat Rev Mol Cell Biol. 2013 May;14(5):269-82. doi: 10.1038/nrm3562. Epub 2013 Apr 18. Nat Rev Mol Cell Biol. 2013. PMID: 23594953 Review.
-
Y-family DNA polymerases in mammalian cells.Cell Mol Life Sci. 2009 Jul;66(14):2363-81. doi: 10.1007/s00018-009-0024-4. Epub 2009 Apr 15. Cell Mol Life Sci. 2009. PMID: 19367366 Free PMC article. Review.
-
Characterization of human translesion DNA synthesis across a UV-induced DNA lesion.Elife. 2016 Oct 22;5:e19788. doi: 10.7554/eLife.19788. Elife. 2016. PMID: 27770570 Free PMC article.
-
Ubiquitin mediates the physical and functional interaction between human DNA polymerases η and ι.Nucleic Acids Res. 2013 Feb 1;41(3):1649-60. doi: 10.1093/nar/gks1277. Epub 2012 Dec 16. Nucleic Acids Res. 2013. PMID: 23248005 Free PMC article.
References
-
- Bienko M., Green C. M., Crosetto N., Rudolf F., Zapart G., Coull B., Kannouche P., Wider G., Peter M., Lehmann A. R., Hofmann K., Dikic I. Ubiquitin-binding domains in translesion synthesis polymerases. Science. 2005;310:1821–1824. - PubMed
-
- Friedberg E. C., Lehmann A. R., Fuchs R. P. Trading places: how do DNA polymerases switch during translesion DNA synthesis? Mol. Cell. 2005;18:499–505. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

