The protein-phosphatome of the human malaria parasite Plasmodium falciparum
- PMID: 18793411
- PMCID: PMC2559854
- DOI: 10.1186/1471-2164-9-412
The protein-phosphatome of the human malaria parasite Plasmodium falciparum
Abstract
Background: Malaria, caused by the parasitic protist Plasmodium falciparum, represents a major public health problem in the developing world. The P. falciparum genome has been sequenced, which provides new opportunities for the identification of novel drug targets. We report an exhaustive analysis of the P. falciparum genomic database (PlasmoDB) aimed at identifying and classifying all protein phosphatases (PP) in this organism.
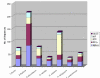
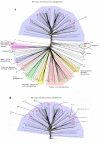
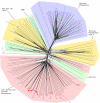
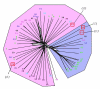
Results: Using a variety of bioinformatics tools, we identified 27 malarial putative PP sequences within the four major established PP families, plus 7 sequences that we predict to dephosphorylate "non-protein" substrates. We constructed phylogenetic trees to position these sequences relative to PPs from other organisms representing all major eukaryotic phyla except Cercozoans (for which no full genome sequence is available). Predominant observations were: (i) P. falciparum possessed the smallest phosphatome of any of the organisms investigated in this study; (ii) no malarial PP clustered with the tyrosine-specific subfamily of the PTP group (iii) a cluster of 7 closely related members of the PPM/PP2C family is present, and (iv) some P. falciparum protein phosphatases are present in clades lacking any human homologue.
Conclusion: The considerable phylogenetic distance between Apicomplexa and other Eukaryotes is reflected by profound divergences between the phosphatome of malaria parasites and those of representative organisms from all major eukaryotic phyla, which might be exploited in the context of efforts for the discovery of novel targets for antimalarial chemotherapy.
Figures


Similar articles
-
Protein kinases of the human malaria parasite Plasmodium falciparum: the kinome of a divergent eukaryote.BMC Genomics. 2004 Oct 12;5:79. doi: 10.1186/1471-2164-5-79. BMC Genomics. 2004. PMID: 15479470 Free PMC article. Review.
-
Genome wide in silico analysis of Plasmodium falciparum phosphatome.BMC Genomics. 2014 Nov 25;15:1024. doi: 10.1186/1471-2164-15-1024. BMC Genomics. 2014. PMID: 25425018 Free PMC article.
-
A genomic glimpse of aminoacyl-tRNA synthetases in malaria parasite Plasmodium falciparum.BMC Genomics. 2009 Dec 31;10:644. doi: 10.1186/1471-2164-10-644. BMC Genomics. 2009. PMID: 20042123 Free PMC article.
-
Protein phosphatase beta, a putative type-2A protein phosphatase from the human malaria parasite Plasmodium falciparum.Eur J Biochem. 1997 Oct 1;249(1):98-106. doi: 10.1111/j.1432-1033.1997.t01-2-00098.x. Eur J Biochem. 1997. PMID: 9363759
-
Plasmodium falciparum serine/threonine phoshoprotein phosphatases (PPP): from housekeeper to the 'holy grail'.Curr Drug Targets. 2008 Nov;9(11):997-1012. doi: 10.2174/138945008786786055. Curr Drug Targets. 2008. PMID: 18991611 Review.
Cited by
-
Atypical mitogen-activated protein kinase phosphatase implicated in regulating transition from pre-S-Phase asexual intraerythrocytic development of Plasmodium falciparum.Eukaryot Cell. 2013 Sep;12(9):1171-8. doi: 10.1128/EC.00028-13. Epub 2013 Jun 28. Eukaryot Cell. 2013. PMID: 23813392 Free PMC article.
-
UIS2: A Unique Phosphatase Required for the Development of Plasmodium Liver Stages.PLoS Pathog. 2016 Jan 6;12(1):e1005370. doi: 10.1371/journal.ppat.1005370. eCollection 2016 Jan. PLoS Pathog. 2016. PMID: 26735921 Free PMC article.
-
Analysis of the Protein phosphotome of Entamoeba histolytica reveals an intricate phosphorylation network.PLoS One. 2013 Nov 13;8(11):e78714. doi: 10.1371/journal.pone.0078714. eCollection 2013. PLoS One. 2013. PMID: 24236039 Free PMC article.
-
Phylogenetic and genetic linkage between novel atypical dual-specificity phosphatases from non-metazoan organisms.Mol Genet Genomics. 2011 Apr;285(4):341-54. doi: 10.1007/s00438-011-0611-6. Epub 2011 Mar 16. Mol Genet Genomics. 2011. PMID: 21409566
-
Genome-wide functional analysis of Plasmodium protein phosphatases reveals key regulators of parasite development and differentiation.Cell Host Microbe. 2014 Jul 9;16(1):128-40. doi: 10.1016/j.chom.2014.05.020. Cell Host Microbe. 2014. PMID: 25011111 Free PMC article.
References
-
- Hanks SK, Quinn AM. Protein kinase catalytic domain sequence database: identification of conserved features of primary structure and classification of family members. Methods Enzymol. 1991;200:38–62. - PubMed
-
- West AH, Stock AM. Histidine kinases and response regulator proteins in two-component signaling systems. Trends Biochem Sci. 2001;26:369–376. - PubMed
-
- Klumpp S, Krieglstein J. Reversible phosphorylation of histidine residues in vertebrate proteins. Biochim Biophys Acta. 2005;1754:291–295. - PubMed
-
- Barford D, Das AK, Egloff MP. The structure and mechanism of protein phosphatases: insights into catalysis and regulation. Annu Rev Biophys Biomol Struct. 1998;27:133–164. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

