Quantum chemical 13C(alpha) chemical shift calculations for protein NMR structure determination, refinement, and validation
- PMID: 18787110
- PMCID: PMC2567219
- DOI: 10.1073/pnas.0807105105
Quantum chemical 13C(alpha) chemical shift calculations for protein NMR structure determination, refinement, and validation
Abstract
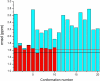
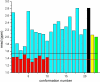
A recently determined set of 20 NMR-derived conformations of a 48-residue all-alpha-helical protein, (PDB ID code 2JVD), is validated here by comparing the observed (13)C(alpha) chemical shifts with those computed at the density functional level of theory. In addition, a recently introduced physics-based method, aimed at determining protein structures by using NOE-derived distance constraints together with observed and computed (13)C(alpha) chemical shifts, was applied to determine a new set of 10 conformations, (Set-bt), as a blind test for the same protein. A cross-validation of these two sets of conformations in terms of the agreement between computed and observed (13)C(alpha) chemical shifts, several stereochemical quality factors, and some NMR quality assessment scores reveals the good quality of both sets of structures. We also carried out an analysis of the agreement between the observed and computed (13)C(alpha) chemical shifts for a slightly longer construct of the protein solved by x-ray crystallography at 2.0-A resolution (PDB ID code 3BHP) with an identical amino acid residue sequence to the 2JVD structure for the first 46 residues. Our results reveal that both of the NMR-derived sets, namely 2JVD and Set-bt, are somewhat better representations of the observed (13)C(alpha) chemical shifts in solution than the 3BHP crystal structure. In addition, the (13)C(alpha)-based validation analysis appears to be more sensitive to subtle structural differences across the three sets of structures than any other NMR quality-assessment scores used here, and, although it is computationally intensive, this analysis has potential value as a standard procedure to determine, refine, and validate protein structures.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
Similar articles
-
Factors affecting the use of 13C(alpha) chemical shifts to determine, refine, and validate protein structures.Proteins. 2008 May 1;71(2):641-54. doi: 10.1002/prot.21726. Proteins. 2008. PMID: 17975838 Free PMC article.
-
Use of 13Calpha chemical shifts in protein structure determination.J Phys Chem B. 2007 Jun 14;111(23):6577-85. doi: 10.1021/jp0683871. Epub 2007 May 22. J Phys Chem B. 2007. PMID: 17516673 Free PMC article.
-
Use of 13C(alpha) chemical shifts for accurate determination of beta-sheet structures in solution.Proc Natl Acad Sci U S A. 2008 Feb 12;105(6):1891-6. doi: 10.1073/pnas.0711022105. Epub 2008 Feb 4. Proc Natl Acad Sci U S A. 2008. PMID: 18250334 Free PMC article.
-
Chemical shift prediction for protein structure calculation and quality assessment using an optimally parameterized force field.Prog Nucl Magn Reson Spectrosc. 2012 Jan;60:1-28. doi: 10.1016/j.pnmrs.2011.05.002. Epub 2011 May 23. Prog Nucl Magn Reson Spectrosc. 2012. PMID: 22293396 Free PMC article. Review.
-
Using quantum chemistry to estimate chemical shifts in biomolecules.Biophys Chem. 2020 Dec;267:106476. doi: 10.1016/j.bpc.2020.106476. Epub 2020 Sep 16. Biophys Chem. 2020. PMID: 33035752 Free PMC article. Review.
Cited by
-
Perspective: Quantum mechanical methods in biochemistry and biophysics.J Chem Phys. 2016 Oct 14;145(14):140901. doi: 10.1063/1.4964410. J Chem Phys. 2016. PMID: 27782516 Free PMC article.
-
Optical signatures of molecular dissymmetry: combining theory with experiments to address stereochemical puzzles.Acc Chem Res. 2009 Jun 16;42(6):809-19. doi: 10.1021/ar8002859. Acc Chem Res. 2009. PMID: 19378940 Free PMC article.
-
Assessing the accuracy of protein structures by quantum mechanical computations of 13C(alpha) chemical shifts.Acc Chem Res. 2009 Oct 20;42(10):1545-53. doi: 10.1021/ar900068s. Acc Chem Res. 2009. PMID: 19572703 Free PMC article.
-
Quality assessment of protein NMR structures.Curr Opin Struct Biol. 2013 Oct;23(5):715-24. doi: 10.1016/j.sbi.2013.08.005. Epub 2013 Sep 21. Curr Opin Struct Biol. 2013. PMID: 24060334 Free PMC article. Review.
-
Automatic 13C chemical shift reference correction for unassigned protein NMR spectra.J Biomol NMR. 2018 Oct;72(1-2):11-28. doi: 10.1007/s10858-018-0202-5. Epub 2018 Aug 10. J Biomol NMR. 2018. PMID: 30097912 Free PMC article.
References
-
- Vila JA, Villegas ME, Baldoni HA, Scheraga HA. Predicting 13Cα chemical shifts for validation of protein structures. J Biomol NMR. 2007;38:221–235. - PubMed
-
- Spera S, Bax A. Empirical correlation between protein backbone conformation and Cα and Cβ 13C nuclear-magnetic-resonance chemical shifts. J Am Chem Soc. 1991;113:5490–5492.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

