Temporal dynamics of cytomegalovirus chromatin assembly in productively infected human cells
- PMID: 18786996
- PMCID: PMC2573275
- DOI: 10.1128/JVI.01218-08
Temporal dynamics of cytomegalovirus chromatin assembly in productively infected human cells
Abstract
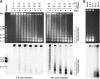
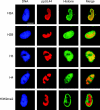
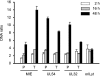
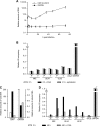
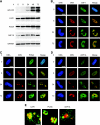
The genomes of herpesviruses, including human cytomegalovirus (CMV), are double-stranded DNA molecules maintained as episomes during infection. The viral DNA lacks histones when encapsidated in the virion. However, it has been found histone associated inside infected cells, implying unidentified chromatin assembly mechanisms. Our results indicate that components of the host cell nucleosome deposition machinery target intranuclear CMV DNA, resulting in stepwise viral-chromatin assembly. CMV genomes undergo limited histone association and nucleosome assembly as early as 30 min after infection via DNA replication-independent mechanisms. Low average viral-genome chromatinization is maintained throughout the early stages of infection. The late phase of infection is characterized by a striking increase in average histone occupancy coupled with the process of viral-DNA replication. While the initial chromatinization affected all analyzed parts of the CMV chromosome, a subset of viral genomic regions, including the major immediate-early promoter, proved to be largely resistant to replication-dependent histone deposition. Finally, our results predict the likely requirement for an unanticipated chromatin disassembly process that enables packaging of histone-free DNA into progeny capsids.
Figures

Similar articles
-
How to control an infectious bead string: nucleosome-based regulation and targeting of herpesvirus chromatin.Rev Med Virol. 2011 May;21(3):154-80. doi: 10.1002/rmv.690. Rev Med Virol. 2011. PMID: 21538665 Review.
-
Chromatinisation of herpesvirus genomes.Rev Med Virol. 2010 Jan;20(1):34-50. doi: 10.1002/rmv.632. Rev Med Virol. 2010. PMID: 19890944 Review.
-
Nucleosome maps of the human cytomegalovirus genome reveal a temporal switch in chromatin organization linked to a major IE protein.Proc Natl Acad Sci U S A. 2013 Aug 6;110(32):13126-31. doi: 10.1073/pnas.1305548110. Epub 2013 Jul 22. Proc Natl Acad Sci U S A. 2013. PMID: 23878222 Free PMC article.
-
Chromatin control of human cytomegalovirus infection.mBio. 2023 Aug 31;14(4):e0032623. doi: 10.1128/mbio.00326-23. Epub 2023 Jul 13. mBio. 2023. PMID: 37439556 Free PMC article. Review.
-
Single cell analysis of RNA-mediated histone H3.3 recruitment to a cytomegalovirus promoter-regulated transcription site.J Biol Chem. 2013 Jul 5;288(27):19882-99. doi: 10.1074/jbc.M113.473181. Epub 2013 May 20. J Biol Chem. 2013. PMID: 23689370 Free PMC article.
Cited by
-
Epigenetic diversity of Kaposi's sarcoma-associated herpesvirus.Nucleic Acids Res. 2013 Mar 1;41(5):2993-3009. doi: 10.1093/nar/gkt033. Epub 2013 Jan 29. Nucleic Acids Res. 2013. PMID: 23361465 Free PMC article.
-
Genome wide nucleosome mapping for HSV-1 shows nucleosomes are deposited at preferred positions during lytic infection.PLoS One. 2015 Feb 24;10(2):e0117471. doi: 10.1371/journal.pone.0117471. eCollection 2015. PLoS One. 2015. PMID: 25710170 Free PMC article.
-
NFκB and Cyclic AMP Response Element Sites Mediate the Valproic Acid and UL138 Responsiveness of the Human Cytomegalovirus Major Immediate Early Enhancer and Promoter.J Virol. 2023 Mar 30;97(3):e0002923. doi: 10.1128/jvi.00029-23. Epub 2023 Mar 1. J Virol. 2023. PMID: 36856444 Free PMC article.
-
Biphasic euchromatin-to-heterochromatin transition on the KSHV genome following de novo infection.PLoS Pathog. 2013;9(12):e1003813. doi: 10.1371/journal.ppat.1003813. Epub 2013 Dec 19. PLoS Pathog. 2013. PMID: 24367262 Free PMC article.
-
Interferon- Stimulation Elicited by the Influenza Virus Is Regulated by the Histone Methylase Dot1L through the RIG-I-TRIM25 Signaling Axis.Cells. 2020 Mar 16;9(3):732. doi: 10.3390/cells9030732. Cells. 2020. PMID: 32188146 Free PMC article.
References
-
- Ahmad, K., and S. Henikoff. 2002. The histone variant H3.3 marks active chromatin by replication-independent nucleosome assembly. Mol. Cell 91191-1200. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials

