Structural insights into intermediate steps in the Sir2 deacetylation reaction
- PMID: 18786399
- PMCID: PMC2590790
- DOI: 10.1016/j.str.2008.05.015
Structural insights into intermediate steps in the Sir2 deacetylation reaction
Abstract
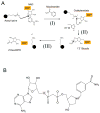
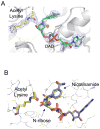
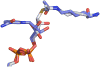
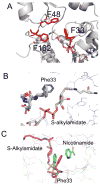
Sirtuin enzymes comprise a unique class of NAD(+)-dependent protein deacetylases. Although structures of many sirtuin complexes have been determined, structural resolution of intermediate chemical steps are needed to understand the deacetylation mechanism. We report crystal structures of the bacterial sirtuin, Sir2Tm, in complex with an S-alkylamidate intermediate, analogous to the naturally occurring O-alkylamidate intermediate, and a Sir2Tm ternary complex containing a dissociated NAD(+) analog and acetylated peptide. The structures and biochemical studies reveal critical roles for the invariant active site histidine in positioning the reaction intermediate, and for a conserved phenylalanine residue in shielding reaction intermediates from base exchange with nicotinamide. The new structural and biochemical studies provide key mechanistic insight into intermediate steps of the Sir2 deacetylation reaction.
Figures

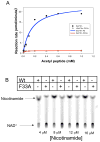

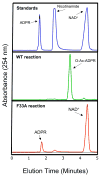
Comment in
-
A SIR-tain acetyl complex is caught by a sulfur trap.Structure. 2008 Sep 10;16(9):1289-92. doi: 10.1016/j.str.2008.08.004. Structure. 2008. PMID: 18786390 No abstract available.
Similar articles
-
Insights into the sirtuin mechanism from ternary complexes containing NAD+ and acetylated peptide.Structure. 2006 Aug;14(8):1231-40. doi: 10.1016/j.str.2006.06.006. Structure. 2006. PMID: 16905097
-
Sir2 regulation by nicotinamide results from switching between base exchange and deacetylation chemistry.Biochemistry. 2003 Aug 12;42(31):9249-56. doi: 10.1021/bi034959l. Biochemistry. 2003. PMID: 12899610
-
Structural basis for nicotinamide cleavage and ADP-ribose transfer by NAD(+)-dependent Sir2 histone/protein deacetylases.Proc Natl Acad Sci U S A. 2004 Jun 8;101(23):8563-8. doi: 10.1073/pnas.0401057101. Epub 2004 May 18. Proc Natl Acad Sci U S A. 2004. PMID: 15150415 Free PMC article.
-
SIR2: the biochemical mechanism of NAD(+)-dependent protein deacetylation and ADP-ribosyl enzyme intermediates.Curr Med Chem. 2004 Apr;11(7):807-26. doi: 10.2174/0929867043455675. Curr Med Chem. 2004. PMID: 15078167 Review.
-
Structure and chemistry of the Sir2 family of NAD+-dependent histone/protein deactylases.Biochem Soc Trans. 2004 Dec;32(Pt 6):904-9. doi: 10.1042/BST0320904. Biochem Soc Trans. 2004. PMID: 15506920 Review.
Cited by
-
SIRT3 substrate specificity determined by peptide arrays and machine learning.ACS Chem Biol. 2011 Feb 18;6(2):146-57. doi: 10.1021/cb100218d. Epub 2010 Nov 1. ACS Chem Biol. 2011. PMID: 20945913 Free PMC article.
-
Sirtuin chemical mechanisms.Biochim Biophys Acta. 2010 Aug;1804(8):1591-603. doi: 10.1016/j.bbapap.2010.01.021. Epub 2010 Feb 2. Biochim Biophys Acta. 2010. PMID: 20132909 Free PMC article. Review.
-
Azalysine analogues as probes for protein lysine deacetylation and demethylation.J Am Chem Soc. 2012 Mar 21;134(11):5138-48. doi: 10.1021/ja209574z. Epub 2012 Mar 12. J Am Chem Soc. 2012. PMID: 22352831 Free PMC article.
-
Sirtuin Deacetylation Mechanism and Catalytic Role of the Dynamic Cofactor Binding Loop.J Phys Chem Lett. 2013 Feb 7;4(3):491-495. doi: 10.1021/jz302015s. J Phys Chem Lett. 2013. PMID: 23585919 Free PMC article.
-
Mechanisms and molecular probes of sirtuins.Chem Biol. 2008 Oct 20;15(10):1002-13. doi: 10.1016/j.chembiol.2008.09.009. Chem Biol. 2008. PMID: 18940661 Free PMC article. Review.
References
-
- Avalos JL, Bever KM, Wolberger C. Mechanism of sirtuin inhibition by nicotinamide: altering the NAD(+) cosubstrate specificity of a Sir2 enzyme. Mol Cell. 2005;17:855–868. - PubMed
-
- Avalos JL, Boeke JD, Wolberger C. Structural basis for the mechanism and regulation of Sir2 enzymes. Mol Cell. 2004;13:639–648. - PubMed
-
- Avalos JL, Celic I, Muhammad S, Cosgrove MS, Boeke JD, Wolberger C. Structure of a Sir2 enzyme bound to an acetylated p53 peptide. Mol Cell. 2002;10:523–535. - PubMed
-
- Bitterman KJ, Anderson RM, Cohen HY, Latorre-Esteves M, Sinclair DA. Inhibition of silencing and accelerated aging by nicotinamide, a putative negative regulator of yeast sir2 and human SIRT1. J Biol Chem. 2002;277:45099–45107. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

