Hierarchical folding mechanism of apomyoglobin revealed by ultra-fast H/D exchange coupled with 2D NMR
- PMID: 18779573
- PMCID: PMC2544544
- DOI: 10.1073/pnas.0804033105
Hierarchical folding mechanism of apomyoglobin revealed by ultra-fast H/D exchange coupled with 2D NMR
Abstract
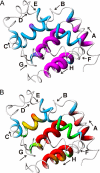
The earliest steps in the folding of proteins are complete on an extremely rapid time scale that is difficult to access experimentally. We have used rapid-mixing quench-flow methods to extend the time resolution of folding studies on apomyoglobin and elucidate the structural and dynamic features of members of the ensemble of intermediate states that are populated on a submillisecond time scale during this process. The picture that emerges is of a continuum of rapidly interconverting states. Even after only 0.4 ms of refolding time a compact state is formed that contains major parts of the A, G, and H helices, which are sufficiently well folded to protect amides from exchange. The B, C, and E helix regions fold more slowly and fluctuate rapidly between open and closed states as they search docking sites on this core; the secondary structure in these regions becomes stabilized as the refolding time is increased from 0.4 to 6 ms. No further stabilization occurs in the A, G, H core at 6 ms of folding time. These studies begin to time-resolve a progression of compact states between the fully unfolded and native folded states and confirm the presence an ensemble of intermediates that interconvert in a hierarchical sequence as the protein searches conformational space on its folding trajectory.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
Similar articles
-
Folding of apomyoglobin: Analysis of transient intermediate structure during refolding using quick hydrogen deuterium exchange and NMR.Proc Jpn Acad Ser B Phys Biol Sci. 2017;93(1):10-27. doi: 10.2183/pjab.93.002. Proc Jpn Acad Ser B Phys Biol Sci. 2017. PMID: 28077807 Free PMC article. Review.
-
NMR structural and dynamic characterization of the acid-unfolded state of apomyoglobin provides insights into the early events in protein folding.Biochemistry. 2001 Mar 27;40(12):3561-71. doi: 10.1021/bi002776i. Biochemistry. 2001. PMID: 11297422
-
Native and non-native secondary structure and dynamics in the pH 4 intermediate of apomyoglobin.Biochemistry. 2000 Mar 21;39(11):2894-901. doi: 10.1021/bi992545f. Biochemistry. 2000. PMID: 10715109
-
How Does Your Protein Fold? Elucidating the Apomyoglobin Folding Pathway.Acc Chem Res. 2017 Jan 17;50(1):105-111. doi: 10.1021/acs.accounts.6b00511. Epub 2016 Dec 29. Acc Chem Res. 2017. PMID: 28032989 Free PMC article.
-
The apomyoglobin folding pathway revisited: structural heterogeneity in the kinetic burst phase intermediate.J Mol Biol. 2002 Sep 20;322(3):483-9. doi: 10.1016/s0022-2836(02)00810-0. J Mol Biol. 2002. PMID: 12225742 Review.
Cited by
-
Transient On- and Off-Pathway Protein Folding Intermediate States Characterized with NMR Relaxation Dispersion.J Phys Chem B. 2022 Nov 24;126(46):9539-9548. doi: 10.1021/acs.jpcb.2c05592. Epub 2022 Nov 10. J Phys Chem B. 2022. PMID: 36354189 Free PMC article.
-
Exploring the Sequence-based Prediction of Folding Initiation Sites in Proteins.Sci Rep. 2017 Aug 18;7(1):8826. doi: 10.1038/s41598-017-08366-3. Sci Rep. 2017. PMID: 28821744 Free PMC article.
-
Evidence for close side-chain packing in an early protein folding intermediate previously assumed to be a molten globule.Proc Natl Acad Sci U S A. 2014 Oct 14;111(41):14746-51. doi: 10.1073/pnas.1410630111. Epub 2014 Sep 25. Proc Natl Acad Sci U S A. 2014. PMID: 25258414 Free PMC article.
-
Folding of apomyoglobin: Analysis of transient intermediate structure during refolding using quick hydrogen deuterium exchange and NMR.Proc Jpn Acad Ser B Phys Biol Sci. 2017;93(1):10-27. doi: 10.2183/pjab.93.002. Proc Jpn Acad Ser B Phys Biol Sci. 2017. PMID: 28077807 Free PMC article. Review.
-
Early Folding Events, Local Interactions, and Conservation of Protein Backbone Rigidity.Biophys J. 2016 Feb 2;110(3):572-583. doi: 10.1016/j.bpj.2015.12.028. Biophys J. 2016. PMID: 26840723 Free PMC article.
References
-
- Uzawa T, et al. Time-resolved small-angle X-ray scattering investigation of the folding dynamics of heme oxygenase: Implication of the scaling relationship for the submillisecond intermediates of protein folding. J Mol Biol. 2006;357:997–1008. - PubMed
-
- Arai M, et al. Microsecond hydrophobic collapse in the folding of Escherichia coli dihydrofolate reductase, an α/β-type protein. J Mol Biol. 2007;368:219–229. - PubMed
-
- Englander SW. Protein folding intermediates and pathways studied by hydrogen exchange. Annu Rev Biophys Biomol Struct. 2000;29:213–238. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

