Inhibition of CED-3 zymogen activation and apoptosis in Caenorhabditis elegans by caspase homolog CSP-3
- PMID: 18776901
- PMCID: PMC2574878
- DOI: 10.1038/nsmb.1488
Inhibition of CED-3 zymogen activation and apoptosis in Caenorhabditis elegans by caspase homolog CSP-3
Abstract
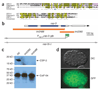
Inhibitor of apoptosis (IAP) proteins have a crucial role in apoptosis, through negative regulation of caspases in species from fruitflies to mammals. In Caenorhabditis elegans, however, no IAP homolog or caspase inhibitor has been identified, calling into question how the cell-killing caspase CED-3 can be negatively regulated. Here we show that inactivation of the C. elegans csp-3 gene, which encodes a protein similar to the small subunit of the CED-3 caspase, causes cells that normally live to undergo apoptosis in a CED-3-dependent manner. Biochemical analysis reveals that CSP-3 associates with the large subunit of the CED-3 zymogen and inhibits zymogen autoactivation. However, CSP-3 does not block CED-3 activation induced by CED-4, nor does it inhibit the activity of the activated CED-3 protease. Therefore CSP-3 uses a previously unreported mechanism to protect cells from apoptosis.
Figures
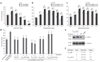



Similar articles
-
Caenorhabditis elegans caspase homolog CSP-2 inhibits CED-3 autoactivation and apoptosis in germ cells.Cell Death Differ. 2009 Oct;16(10):1385-94. doi: 10.1038/cdd.2009.88. Epub 2009 Jul 3. Cell Death Differ. 2009. PMID: 19575016 Free PMC article.
-
Both the caspase CSP-1 and a caspase-independent pathway promote programmed cell death in parallel to the canonical pathway for apoptosis in Caenorhabditis elegans.PLoS Genet. 2013;9(3):e1003341. doi: 10.1371/journal.pgen.1003341. Epub 2013 Mar 7. PLoS Genet. 2013. PMID: 23505386 Free PMC article.
-
Identification of multiple Caenorhabditis elegans caspases and their potential roles in proteolytic cascades.J Biol Chem. 1998 Dec 25;273(52):35109-17. doi: 10.1074/jbc.273.52.35109. J Biol Chem. 1998. PMID: 9857046
-
2:1 Stoichiometry of the CED-4-CED-9 complex and the tetrameric CED-4: insights into the regulation of CED-3 activation.Cell Cycle. 2006 Jan;5(1):31-4. doi: 10.4161/cc.5.1.2263. Epub 2006 Jan 18. Cell Cycle. 2006. PMID: 16294007 Review.
-
Apoptosis: a process with a (beta)NAC for complexity.Cell. 2003 Sep 19;114(6):659-61. doi: 10.1016/s0092-8674(03)00717-7. Cell. 2003. PMID: 14505566 Review.
Cited by
-
C. elegans EIF-3.K promotes programmed cell death through CED-3 caspase.PLoS One. 2012;7(5):e36584. doi: 10.1371/journal.pone.0036584. Epub 2012 May 9. PLoS One. 2012. PMID: 22590572 Free PMC article.
-
Regulation of CED-3 caspase localization and activation by C. elegans nuclear-membrane protein NPP-14.Nat Struct Mol Biol. 2016 Nov;23(11):958-964. doi: 10.1038/nsmb.3308. Epub 2016 Oct 10. Nat Struct Mol Biol. 2016. PMID: 27723735 Free PMC article.
-
A caspase-like decoy molecule enhances the activity of a paralogous caspase in the yellow fever mosquito, Aedes aegypti.Insect Biochem Mol Biol. 2010 Jul;40(7):516-23. doi: 10.1016/j.ibmb.2010.04.011. Epub 2010 Apr 24. Insect Biochem Mol Biol. 2010. PMID: 20417712 Free PMC article.
-
Post-transcriptional control of executioner caspases by RNA-binding proteins.Genes Dev. 2016 Oct 1;30(19):2213-2225. doi: 10.1101/gad.285726.116. Genes Dev. 2016. PMID: 27798844 Free PMC article.
-
Programmed cell death and clearance of cell corpses in Caenorhabditis elegans.Cell Mol Life Sci. 2016 Jun;73(11-12):2221-36. doi: 10.1007/s00018-016-2196-z. Epub 2016 Apr 5. Cell Mol Life Sci. 2016. PMID: 27048817 Free PMC article. Review.
References
-
- Steller H. Mechanisms and genes of cellular suicide. Science. 1995;267:1445–1449. - PubMed
-
- Thornberry NA, Lazebnik Y. Caspases: enemies within. Science. 1998;281:1312–1316. - PubMed
-
- Boatright KM, Salvesen GS. Mechanisms of caspase activation. Curr. Opin. Cell Biol. 2003;15:725–731. - PubMed
-
- Shi Y. Mechanisms of caspase activation and inhibition during apoptosis. Mol. Cell. 2002;9:459–470. - PubMed
-
- Budihardjo I, Oliver H, Lutter M, Luo X, Wang X. Biochemical pathways of caspase activation during apoptosis. Annu. Rev. Cell Dev. Biol. 1999;15:269–290. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

