Oxidative demethylation of 3-methylthymine and 3-methyluracil in single-stranded DNA and RNA by mouse and human FTO
- PMID: 18775698
- PMCID: PMC2577709
- DOI: 10.1016/j.febslet.2008.08.019
Oxidative demethylation of 3-methylthymine and 3-methyluracil in single-stranded DNA and RNA by mouse and human FTO
Abstract
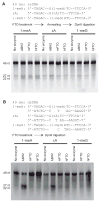
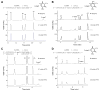
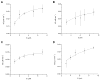
The human obesity susceptibility gene, FTO, encodes a protein that is homologous to the DNA repair AlkB protein. The AlkB family proteins utilize iron(II), alpha-ketoglutarate (alpha-KG) and dioxygen to perform oxidative repair of alkylated nucleobases in DNA and RNA. We demonstrate here the oxidative demethylation of 3-methylthymine (3-meT) in single-stranded DNA (ssDNA) and 3-methyluracil (3-meU) in single-stranded RNA (ssRNA) by recombinant human FTO protein in vitro. Both human and mouse FTO proteins preferentially repair 3-meT in ssDNA over other base lesions tested. They showed negligible activities against 3-meT in double-stranded DNA (dsDNA). In addition, these two proteins can catalyze the demethylation of 3-meU in ssRNA with a slightly higher efficiency over that of 3-meT in ssDNA, suggesting that methylated RNAs are the preferred substrates for FTO.
Figures

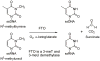
Similar articles
-
A non-heme iron-mediated chemical demethylation in DNA and RNA.Acc Chem Res. 2009 Apr 21;42(4):519-29. doi: 10.1021/ar800178j. Acc Chem Res. 2009. PMID: 19852088 Free PMC article.
-
Crystal structure of the FTO protein reveals basis for its substrate specificity.Nature. 2010 Apr 22;464(7292):1205-9. doi: 10.1038/nature08921. Epub 2010 Apr 7. Nature. 2010. PMID: 20376003
-
Structural insights into FTO's catalytic mechanism for the demethylation of multiple RNA substrates.Proc Natl Acad Sci U S A. 2019 Feb 19;116(8):2919-2924. doi: 10.1073/pnas.1820574116. Epub 2019 Feb 4. Proc Natl Acad Sci U S A. 2019. PMID: 30718435 Free PMC article.
-
The AlkB Family of Fe(II)/α-Ketoglutarate-dependent Dioxygenases: Repairing Nucleic Acid Alkylation Damage and Beyond.J Biol Chem. 2015 Aug 21;290(34):20734-20742. doi: 10.1074/jbc.R115.656462. Epub 2015 Jul 7. J Biol Chem. 2015. PMID: 26152727 Free PMC article. Review.
-
ALKBHs-facilitated RNA modifications and de-modifications.DNA Repair (Amst). 2016 Aug;44:87-91. doi: 10.1016/j.dnarep.2016.05.026. Epub 2016 May 17. DNA Repair (Amst). 2016. PMID: 27237585 Free PMC article. Review.
Cited by
-
tRNA expression and modification landscapes, and their dynamics during zebrafish embryo development.Nucleic Acids Res. 2024 Sep 23;52(17):10575-10594. doi: 10.1093/nar/gkae595. Nucleic Acids Res. 2024. PMID: 38989621 Free PMC article.
-
Emerging Role of m6 A Methylome in Brain Development: Implications for Neurological Disorders and Potential Treatment.Front Cell Dev Biol. 2021 May 19;9:656849. doi: 10.3389/fcell.2021.656849. eCollection 2021. Front Cell Dev Biol. 2021. PMID: 34095121 Free PMC article. Review.
-
Recent Advances of RNA m6A Modifications in Cancer Immunoediting and Immunotherapy.Cancer Treat Res. 2023;190:49-94. doi: 10.1007/978-3-031-45654-1_3. Cancer Treat Res. 2023. PMID: 38112999
-
The Biogenesis and Precise Control of RNA m6A Methylation.Trends Genet. 2020 Jan;36(1):44-52. doi: 10.1016/j.tig.2019.10.011. Epub 2019 Dec 4. Trends Genet. 2020. PMID: 31810533 Free PMC article. Review.
-
Evaluation of FTO rs9939609 and MC4R rs17782313 Polymorphisms as Prognostic Biomarkers of Obesity: A Population-based Cross-sectional Study.Oman Med J. 2019 Jan;34(1):56-62. doi: 10.5001/omj.2019.09. Oman Med J. 2019. PMID: 30671185 Free PMC article.
References
-
- Sedgwick B, Bates PA, Paik J, Jacobs SC, Lindahl T. Repair of alkylated DNA: recent advances. DNA Repair. 2007;6:429–442. - PubMed
-
- Yu B, Edstrom EC, Benach J, Hamuro Y, Weber PC, Gibney BR, Hunt JF. Crystal structures of catalytic complexes of the oxidative DNA/RNA repair enzyme AlkB. Nature. 2006;439:879–884. - PubMed
-
- Aas PA, Otterlei M, Falnes PØ, Vågbø CB, Skorpen F, Akbari M, Sundheim O, Bjørås M, Slupphaug G, Seeberg E, Krokan HE. Human and bacterial oxidative demethylases repair alkylation damage in both RNA and DNA. Nature. 2003;421:859–863. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

