Competition and collaboration: GATA-3, PU.1, and Notch signaling in early T-cell fate determination
- PMID: 18768329
- PMCID: PMC2634812
- DOI: 10.1016/j.smim.2008.07.006
Competition and collaboration: GATA-3, PU.1, and Notch signaling in early T-cell fate determination
Abstract
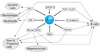
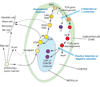
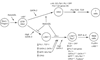
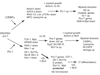
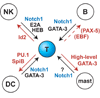
T-cell precursors remain developmentally plastic for multiple cell generations after entering the thymus, preserving access to developmental alternatives of macrophage, dendritic-cell, and even mast-cell fates. The underlying regulatory basis of this plasticity is that early T-cell differentiation depends on transcription factors which can also promote alternative developmental programs. Interfactor competition, together with environmental signals, keep these diversions under control. Here the pathways leading to several lineage alternatives for early pro-T-cells are reviewed, with close focus on the mechanisms of action of three vital factors, GATA-3, PU.1, and Notch-Delta signals, whose counterbalance appears to be essential for T-cell specification.
Figures
Similar articles
-
Architecture of a lymphomyeloid developmental switch controlled by PU.1, Notch and Gata3.Development. 2013 Mar;140(6):1207-19. doi: 10.1242/dev.088559. Development. 2013. PMID: 23444353 Free PMC article.
-
A gene regulatory network armature for T lymphocyte specification.Proc Natl Acad Sci U S A. 2008 Dec 23;105(51):20100-5. doi: 10.1073/pnas.0806501105. Epub 2008 Dec 22. Proc Natl Acad Sci U S A. 2008. PMID: 19104054 Free PMC article.
-
Notch/Delta signaling constrains reengineering of pro-T cells by PU.1.Proc Natl Acad Sci U S A. 2006 Aug 8;103(32):11993-8. doi: 10.1073/pnas.0601188103. Epub 2006 Jul 31. Proc Natl Acad Sci U S A. 2006. PMID: 16880393 Free PMC article.
-
Transcriptional establishment of cell-type identity: dynamics and causal mechanisms of T-cell lineage commitment.Cold Spring Harb Symp Quant Biol. 2013;78:31-41. doi: 10.1101/sqb.2013.78.020271. Epub 2013 Oct 17. Cold Spring Harb Symp Quant Biol. 2013. PMID: 24135716 Free PMC article. Review.
-
Forging T-Lymphocyte Identity: Intersecting Networks of Transcriptional Control.Adv Immunol. 2016;129:109-74. doi: 10.1016/bs.ai.2015.09.002. Epub 2015 Oct 26. Adv Immunol. 2016. PMID: 26791859 Free PMC article. Review.
Cited by
-
Bcl11b-A Critical Neurodevelopmental Transcription Factor-Roles in Health and Disease.Front Cell Neurosci. 2017 Mar 29;11:89. doi: 10.3389/fncel.2017.00089. eCollection 2017. Front Cell Neurosci. 2017. PMID: 28424591 Free PMC article. Review.
-
The signal transducers STAT5 and STAT3 control expression of Id2 and E2-2 during dendritic cell development.Blood. 2012 Nov 22;120(22):4363-73. doi: 10.1182/blood-2012-07-441311. Epub 2012 Oct 1. Blood. 2012. PMID: 23033267 Free PMC article.
-
GATA2-Mediated Transcriptional Activation of Notch3 Promotes Pancreatic Cancer Liver Metastasis.Mol Cells. 2022 May 31;45(5):329-342. doi: 10.14348/molcells.2022.2176. Mol Cells. 2022. PMID: 35534193 Free PMC article.
-
Mechanisms regulating dendritic cell specification and development.Immunol Rev. 2010 Nov;238(1):76-92. doi: 10.1111/j.1600-065X.2010.00949.x. Immunol Rev. 2010. PMID: 20969586 Free PMC article. Review.
-
Gata3/Ruvbl2 complex regulates T helper 2 cell proliferation via repression of Cdkn2c expression.Proc Natl Acad Sci U S A. 2013 Nov 12;110(46):18626-31. doi: 10.1073/pnas.1311100110. Epub 2013 Oct 28. Proc Natl Acad Sci U S A. 2013. PMID: 24167278 Free PMC article.
References
-
- Laiosa CV, Stadtfeld M, Graf T. Determinants of lymphoid-myeloid lineage diversification. Annu Rev Immunol. 2006;24:705–738. - PubMed
-
- Rhodes J, Hagen A, Hsu K, Deng M, Liu TX, Look AT, et al. Interplay of pu.1 and gata1 determines myelo-erythroid progenitor cell fate in zebrafish. Dev Cell. 2005;8:97–108. - PubMed
-
- Graf T. Transcription factors that induce commitment of multipotent hematopoietic progenitors: lessons from the MEP system. In: Zon LI, editor. Hematopoiesis: a Developmental Approach. New York: Oxford University Press; 2001. pp. 355–362.
-
- Nerlov C, Querfurth E, Kulessa H, Graf T. GATA-1 interacts with the myeloid PU.1 transcription factor and represses PU.1-dependent transcription. Blood. 2000;95:2543–2551. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

