Structure of the guide-strand-containing argonaute silencing complex
- PMID: 18754009
- PMCID: PMC4689319
- DOI: 10.1038/nature07315
Structure of the guide-strand-containing argonaute silencing complex
Abstract
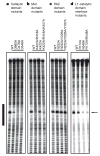
The slicer activity of the RNA-induced silencing complex is associated with argonaute, the RNase H-like PIWI domain of which catalyses guide-strand-mediated sequence-specific cleavage of target messenger RNA. Here we report on the crystal structure of Thermus thermophilus argonaute bound to a 5'-phosphorylated 21-base DNA guide strand, thereby identifying the nucleic-acid-binding channel positioned between the PAZ- and PIWI-containing lobes, as well as the pivot-like conformational changes associated with complex formation. The bound guide strand is anchored at both of its ends, with the solvent-exposed Watson-Crick edges of stacked bases 2 to 6 positioned for nucleation with the mRNA target, whereas two critically positioned arginines lock bases 10 and 11 at the cleavage site into an unanticipated orthogonal alignment. Biochemical studies indicate that key amino acid residues at the active site and those lining the 5'-phosphate-binding pocket made up of the Mid domain are critical for cleavage activity, whereas alterations of residues lining the 2-nucleotide 3'-end-binding pocket made up of the PAZ domain show little effect.
Figures
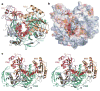
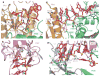
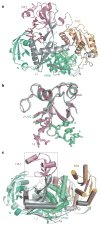
Similar articles
-
Structure of an argonaute silencing complex with a seed-containing guide DNA and target RNA duplex.Nature. 2008 Dec 18;456(7224):921-6. doi: 10.1038/nature07666. Nature. 2008. PMID: 19092929 Free PMC article.
-
Nucleation, propagation and cleavage of target RNAs in Ago silencing complexes.Nature. 2009 Oct 8;461(7265):754-61. doi: 10.1038/nature08434. Nature. 2009. PMID: 19812667 Free PMC article.
-
Structure/cleavage-based insights into helical perturbations at bulge sites within T. thermophilus Argonaute silencing complexes.Nucleic Acids Res. 2017 Sep 6;45(15):9149-9163. doi: 10.1093/nar/gkx547. Nucleic Acids Res. 2017. PMID: 28911094 Free PMC article.
-
Crystal structure of A. aeolicus argonaute, a site-specific DNA-guided endoribonuclease, provides insights into RISC-mediated mRNA cleavage.Mol Cell. 2005 Aug 5;19(3):405-19. doi: 10.1016/j.molcel.2005.07.011. Mol Cell. 2005. PMID: 16061186 Free PMC article.
-
Understanding the core of RNA interference: The dynamic aspects of Argonaute-mediated processes.Prog Biophys Mol Biol. 2017 Sep;128:39-46. doi: 10.1016/j.pbiomolbio.2016.09.008. Epub 2016 Sep 30. Prog Biophys Mol Biol. 2017. PMID: 27697475 Review.
Cited by
-
Comparative genomics of defense systems in archaea and bacteria.Nucleic Acids Res. 2013 Apr;41(8):4360-77. doi: 10.1093/nar/gkt157. Epub 2013 Mar 6. Nucleic Acids Res. 2013. PMID: 23470997 Free PMC article. Review.
-
Alternative RISC assembly: binding and repression of microRNA-mRNA duplexes by human Ago proteins.RNA. 2012 Nov;18(11):2041-55. doi: 10.1261/rna.035675.112. Epub 2012 Sep 27. RNA. 2012. PMID: 23019594 Free PMC article.
-
miRNA-like duplexes as RNAi triggers with improved specificity.Front Genet. 2012 Jul 12;3:127. doi: 10.3389/fgene.2012.00127. eCollection 2012. Front Genet. 2012. PMID: 22807929 Free PMC article.
-
The structure of human argonaute-2 in complex with miR-20a.Cell. 2012 Jul 6;150(1):100-10. doi: 10.1016/j.cell.2012.05.017. Epub 2012 Jun 7. Cell. 2012. PMID: 22682761 Free PMC article.
-
Regulation of microRNA function in animals.Nat Rev Mol Cell Biol. 2019 Jan;20(1):21-37. doi: 10.1038/s41580-018-0045-7. Nat Rev Mol Cell Biol. 2019. PMID: 30108335 Free PMC article. Review.
References
-
- Baulcombe D. RNA silencing in plants. Nature. 2004;431:356–363. - PubMed
-
- Filipowicz W. The nuts and bolts of the RISC machine. Cell. 2005;122:17–20. - PubMed
-
- Rana TM. Illuminating the silence: understanding the structure and function of small RNAs. Nature Rev Mol Cell Biol. 2007;8:23–36. - PubMed
-
- Kim DH, Rossi JJ. Strategies for silencing human disease using RNA interference. Nature Rev Genet. 2007;8:173–184. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

