Lysine acetylation: codified crosstalk with other posttranslational modifications
- PMID: 18722172
- PMCID: PMC2551738
- DOI: 10.1016/j.molcel.2008.07.002
Lysine acetylation: codified crosstalk with other posttranslational modifications
Abstract
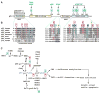
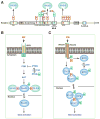
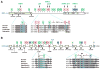
Lysine acetylation has emerged as a major posttranslational modification for histones. Crossregulation between this and other modifications is crucial in modulating chromatin-based transcriptional control and shaping inheritable epigenetic programs. In addition to histones, many other nuclear proteins and various cytoplasmic regulators are subject to lysine acetylation. This review focuses on recent findings pertinent to acetylation of nonhistone proteins and emphasizes how this modification might crosstalk with phosphorylation, methylation, ubiquitination, sumoylation, and others to form code-like multisite modification programs for dynamic control of cellular signaling under diverse conditions.
Figures



Similar articles
-
Crosstalk between CARM1 methylation and CBP acetylation on histone H3.Curr Biol. 2002 Dec 23;12(24):2090-7. doi: 10.1016/s0960-9822(02)01387-8. Curr Biol. 2002. PMID: 12498683
-
Lysine acetylation: enzymes, bromodomains and links to different diseases.Essays Biochem. 2012;52:1-12. doi: 10.1042/bse0520001. Essays Biochem. 2012. PMID: 22708559 Review.
-
Lysine methylation: beyond histones.Acta Biochim Biophys Sin (Shanghai). 2012 Jan;44(1):14-27. doi: 10.1093/abbs/gmr100. Acta Biochim Biophys Sin (Shanghai). 2012. PMID: 22194010 Review.
-
Aberrant lysine acetylation in tumorigenesis: Implications in the development of therapeutics.Pharmacol Ther. 2016 Jun;162:98-119. doi: 10.1016/j.pharmthera.2016.01.011. Epub 2016 Jan 22. Pharmacol Ther. 2016. PMID: 26808162 Review.
-
Beyond histones - the expanding roles of protein lysine methylation.FEBS J. 2017 Sep;284(17):2732-2744. doi: 10.1111/febs.14056. Epub 2017 May 2. FEBS J. 2017. PMID: 28294537 Review.
Cited by
-
Novel brd4 inhibitors with a unique scaffold exhibit antitumor effects.Oncol Lett. 2021 Jun;21(6):473. doi: 10.3892/ol.2021.12734. Epub 2021 Apr 14. Oncol Lett. 2021. PMID: 33907583 Free PMC article.
-
Methylation as a key regulator of Tau aggregation and neuronal health in Alzheimer's disease.Cell Commun Signal. 2021 May 7;19(1):51. doi: 10.1186/s12964-021-00732-z. Cell Commun Signal. 2021. PMID: 33962636 Free PMC article. Review.
-
Reactivation of latent HIV by histone deacetylase inhibitors.Trends Microbiol. 2013 Jun;21(6):277-85. doi: 10.1016/j.tim.2013.02.005. Epub 2013 Mar 18. Trends Microbiol. 2013. PMID: 23517573 Free PMC article. Review.
-
Targeting Histone Modifications in Bone and Lung Metastatic Cancers.Curr Osteoporos Rep. 2021 Jun;19(3):230-246. doi: 10.1007/s11914-021-00670-2. Epub 2021 Mar 15. Curr Osteoporos Rep. 2021. PMID: 33721181 Free PMC article. Review.
-
Cortactin tyrosine phosphorylation promotes its deacetylation and inhibits cell spreading.PLoS One. 2012;7(3):e33662. doi: 10.1371/journal.pone.0033662. Epub 2012 Mar 30. PLoS One. 2012. PMID: 22479425 Free PMC article.
References
-
- Allis CD, Berger SL, Cote J, Dent S, Jenuwien T, Kouzarides T, Pillus L, Reinberg D, Shi Y, Shiekhattar R, et al. New nomenclature for chromatin-modifying enzymes. Cell. 2007;131:633–636. - PubMed
-
- Appella E, Anderson CW. Signaling to p53: breaking the posttranslational modification code. Pathol Biol (Paris) 2000;48:227–245. - PubMed
-
- Bali P, Pranpat M, Bradner J, Balasis M, Fiskus W, Guo F, Rocha K, Kumaraswamy S, Boyapalle S, Atadja P, et al. Inhibition of histone deacetylase 6 acetylates and disrupts the chaperone function of heat shock protein 90: a novel basis for antileukemia activity of histone deacetylase inhibitors. J Biol Chem. 2005;280:26729–26734. - PubMed
-
- Berger SL. The complex language of chromatin regulation during transcription. Nature. 2007;447:407–412. - PubMed
-
- Bhaumik SR, Smith E, Shilatifard A. Covalent modifications of histones during development and disease pathogenesis. Nat Struct Mol Biol. 2007;14:1008–1016. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

