Quantitative analysis of redox-sensitive proteome with DIGE and ICAT
- PMID: 18707151
- PMCID: PMC2577071
- DOI: 10.1021/pr800233r
Quantitative analysis of redox-sensitive proteome with DIGE and ICAT
Abstract
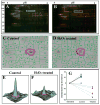
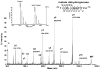
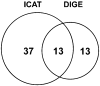
Oxidative modifications of protein thiols are important mechanisms for regulating protein functions. The present study aimed to compare the relative effectiveness of two thiol-specific quantitative proteomic techniques, difference gel electrophoresis (DIGE) and isotope coded affinity tag (ICAT), for the discovery of redox-sensitive proteins in heart tissues. We found that these two methods were largely complementary; each could be used to reveal a set of unique redox-sensitive proteins. Some of these proteins are low-abundant signaling proteins and membrane proteins. From DIGE analysis, we found that both NF-kappaB-repressing protein and epoxide hydrolase were sensitive to H 2O 2 oxidation. In ICAT analysis, we found that specific cysteines within sacroplasmic endoplamic reticulum calcium ATPase 2 and voltage-dependent anion-selective channel protein 1 were sensitive to H 2O 2 oxidation. From these analyses, we conclude that both methods should be employed for proteome-wide studies, to maximize the possibility of identifying proteins containing redox-sensitive cysteinyl thiols in complex biological systems.
Figures

Similar articles
-
Isotope-coded affinity tag (ICAT) approach to redox proteomics: identification and quantitation of oxidant-sensitive cysteine thiols in complex protein mixtures.J Proteome Res. 2004 Nov-Dec;3(6):1228-33. doi: 10.1021/pr049887e. J Proteome Res. 2004. PMID: 15595732
-
Isotope-coded affinity tag approach to identify and quantify oxidant-sensitive protein thiols.Mol Cell Proteomics. 2004 Mar;3(3):273-8. doi: 10.1074/mcp.T300011-MCP200. Epub 2004 Jan 15. Mol Cell Proteomics. 2004. PMID: 14726493
-
The oxidized thiol proteome in fission yeast--optimization of an ICAT-based method to identify H2O2-oxidized proteins.J Proteomics. 2011 Oct 19;74(11):2476-86. doi: 10.1016/j.jprot.2011.05.030. Epub 2011 Jun 6. J Proteomics. 2011. PMID: 21672643
-
Concepts and approaches towards understanding the cellular redox proteome.Plant Biol (Stuttg). 2006 Jul;8(4):407-18. doi: 10.1055/s-2006-923961. Plant Biol (Stuttg). 2006. PMID: 16906481 Review.
-
Proteomic Characterization of Reversible Thiol Oxidations in Proteomes and Proteins.Antioxid Redox Signal. 2017 Mar 1;26(7):329-344. doi: 10.1089/ars.2016.6720. Epub 2016 May 20. Antioxid Redox Signal. 2017. PMID: 27089838 Review.
Cited by
-
Functional proteomics approaches for the identification of transnitrosylase and denitrosylase targets.Methods. 2013 Aug 1;62(2):151-60. doi: 10.1016/j.ymeth.2013.02.002. Epub 2013 Feb 18. Methods. 2013. PMID: 23428400 Free PMC article.
-
Quantitative redox proteomics revealed molecular mechanisms of salt tolerance in the roots of sugar beet monomeric addition line M14.Bot Stud. 2022 Mar 5;63(1):5. doi: 10.1186/s40529-022-00337-w. Bot Stud. 2022. PMID: 35247135 Free PMC article.
-
Characterization of thiol-based redox modifications of Brassica napusSNF1-related protein kinase 2.6-2C.FEBS Open Bio. 2018 Mar 5;8(4):628-645. doi: 10.1002/2211-5463.12401. eCollection 2018 Apr. FEBS Open Bio. 2018. PMID: 29632815 Free PMC article.
-
Chasing cysteine oxidative modifications: proteomic tools for characterizing cysteine redox status.Circ Cardiovasc Genet. 2012 Oct 1;5(5):591. doi: 10.1161/CIRCGENETICS.111.961425. Circ Cardiovasc Genet. 2012. PMID: 23074338 Free PMC article.
-
Regulatory control or oxidative damage? Proteomic approaches to interrogate the role of cysteine oxidation status in biological processes.Mol Cell Proteomics. 2012 Apr;11(4):R111.013037. doi: 10.1074/mcp.R111.013037. Epub 2011 Dec 8. Mol Cell Proteomics. 2012. PMID: 22159599 Free PMC article. Review.
References
-
- Ghezzi P, Bonetto V. Redox proteomics: Identification of oxidatively, modified proteins. Proteomics. 2003;3(7):1145–53. - PubMed
-
- Di Simplicio P, Franconi F, Frosali S, Di Giuseppe D. Thiolation and nitrosation of cysteines in biological fluids and cells. Amino Acids. 2003;25(3–4):323–39. - PubMed
-
- Zheng M, Aslund F, Storz G. Activation of the OxyR transcription factor by reversible disulfide bond formation. Science. 1998;279(5357):1718–21. - PubMed
-
- Abate C, Patel L, Rauscher FJ, Curran T. Redox regulation of fos and jun DNA-binding activity in vitro. Science. 1990;249(4973):1157–61. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

