Selective constraints in experimentally defined primate regulatory regions
- PMID: 18704158
- PMCID: PMC2490716
- DOI: 10.1371/journal.pgen.1000157
Selective constraints in experimentally defined primate regulatory regions
Abstract
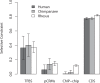
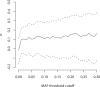
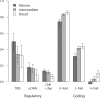
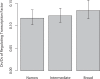
Changes in gene regulation may be important in evolution. However, the evolutionary properties of regulatory mutations are currently poorly understood. This is partly the result of an incomplete annotation of functional regulatory DNA in many species. For example, transcription factor binding sites (TFBSs), a major component of eukaryotic regulatory architecture, are typically short, degenerate, and therefore difficult to differentiate from randomly occurring, nonfunctional sequences. Furthermore, although sites such as TFBSs can be computationally predicted using evolutionary conservation as a criterion, estimates of the true level of selective constraint (defined as the fraction of strongly deleterious mutations occurring at a locus) in regulatory regions will, by definition, be upwardly biased in datasets that are a priori evolutionarily conserved. Here we investigate the fitness effects of regulatory mutations using two complementary datasets of human TFBSs that are likely to be relatively free of ascertainment bias with respect to evolutionary conservation but, importantly, are supported by experimental data. The first is a collection of almost >2,100 human TFBSs drawn from the literature in the TRANSFAC database, and the second is derived from several recent high-throughput chromatin immunoprecipitation coupled with genomic microarray (ChIP-chip) analyses. We also define a set of putative cis-regulatory modules (pCRMs) by spatially clustering multiple TFBSs that regulate the same gene. We find that a relatively high proportion ( approximately 37%) of mutations at TFBSs are strongly deleterious, similar to that at a 2-fold degenerate protein-coding site. However, constraint is significantly reduced in human and chimpanzee pCRMS and ChIP-chip sequences, relative to macaques. We estimate that the fraction of regulatory mutations that have been driven to fixation by positive selection in humans is not significantly different from zero. We also find that the level of selective constraint in our TFBSs, pCRMs, and ChIP-chip sequences is negatively correlated with the expression breadth of the regulated gene, whereas the opposite relationship holds at that gene's nonsynonymous and synonymous sites. Finally, we find that the rate of protein evolution in a transcription factor appears to be positively correlated with the breadth of expression of the gene it regulates. Our study suggests that strongly deleterious regulatory mutations are considerably more likely (1.6-fold) to occur in tissue-specific than in housekeeping genes, implying that there is a fitness cost to increasing "complexity" of gene expression.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures
Similar articles
-
De novo prediction of cis-regulatory elements and modules through integrative analysis of a large number of ChIP datasets.BMC Genomics. 2014 Dec 2;15:1047. doi: 10.1186/1471-2164-15-1047. BMC Genomics. 2014. PMID: 25442502 Free PMC article.
-
Mammalian evolution of human cis-regulatory elements and transcription factor binding sites.Science. 2023 Apr 28;380(6643):eabn7930. doi: 10.1126/science.abn7930. Epub 2023 Apr 28. Science. 2023. PMID: 37104580
-
Genome-wide prediction of transcriptional regulatory elements of human promoters using gene expression and promoter analysis data.BMC Bioinformatics. 2006 Jul 4;7:330. doi: 10.1186/1471-2105-7-330. BMC Bioinformatics. 2006. PMID: 16817975 Free PMC article.
-
Nonadaptive processes in primate and human evolution.Am J Phys Anthropol. 2010;143 Suppl 51:13-45. doi: 10.1002/ajpa.21439. Am J Phys Anthropol. 2010. PMID: 21086525 Review.
-
Identification of altered cis-regulatory elements in human disease.Trends Genet. 2015 Feb;31(2):67-76. doi: 10.1016/j.tig.2014.12.003. Epub 2015 Jan 27. Trends Genet. 2015. PMID: 25637093 Review.
Cited by
-
A selection index for gene expression evolution and its application to the divergence between humans and chimpanzees.PLoS One. 2012;7(4):e34935. doi: 10.1371/journal.pone.0034935. Epub 2012 Apr 18. PLoS One. 2012. PMID: 22529958 Free PMC article.
-
Evolutionary processes acting on candidate cis-regulatory regions in humans inferred from patterns of polymorphism and divergence.PLoS Genet. 2009 Aug;5(8):e1000592. doi: 10.1371/journal.pgen.1000592. Epub 2009 Aug 7. PLoS Genet. 2009. PMID: 19662163 Free PMC article.
-
Evolutionary dynamics of coding and non-coding transcriptomes.Nat Rev Genet. 2014 Nov;15(11):734-48. doi: 10.1038/nrg3802. Epub 2014 Oct 9. Nat Rev Genet. 2014. PMID: 25297727 Review.
-
Genome-wide inference of natural selection on human transcription factor binding sites.Nat Genet. 2013 Jul;45(7):723-9. doi: 10.1038/ng.2658. Epub 2013 Jun 9. Nat Genet. 2013. PMID: 23749186 Free PMC article.
-
Evolution of neuronal and endothelial transcriptomes in primates.Genome Biol Evol. 2010 Jul 12;2:284-92. doi: 10.1093/gbe/evq018. Genome Biol Evol. 2010. PMID: 20624733 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

