Posttranscriptional regulation of II10 gene expression allows natural killer cells to express immunoregulatory function
- PMID: 18701085
- PMCID: PMC2656759
- DOI: 10.1016/j.immuni.2008.06.012
Posttranscriptional regulation of II10 gene expression allows natural killer cells to express immunoregulatory function
Abstract
Natural killer (NK) cells play a well-recognized role in early pathogen containment and in shaping acquired cell-mediated immunity. However, indirect evidence in humans and experimental models has suggested that NK cells also play negative regulatory roles during chronic disease. To formally test this hypothesis, we employed a well-defined experimental model of visceral leishmaniasis. Our data demonstrated that NKp46(+)CD49b(+)CD3(-) NK cells were recruited to the spleen and into hepatic granulomas, where they inhibited host protective immunity in an interleukin-10 (IL-10)-dependent manner. Although IL-10 mRNA could be detected in activated NK cells 24 hr after infection, the inhibitory function of NK cells was only acquired later during infection, coincident with increased IL-10 mRNA stability and an enhanced capacity to secrete IL-10 protein. Our data support a growing body of literature that implicates NK cells as negative regulators of cell-mediated immunity and suggest that NK cells, like CD4(+) T helper 1 cells, may acquire immunoregulatory functions as a consequence of extensive activation.
Figures

 ) and CD8+CD3+ (
) and CD8+CD3+ ( ) T cells, B220+ B cells (
) T cells, B220+ B cells ( ), CD49b+ NK cells (
), CD49b+ NK cells ( ), CD11chi DC (
), CD11chi DC ( ) and all residual cells (including macrophages and stromal cells;
) and all residual cells (including macrophages and stromal cells;  ) were sorted to >99% purity and then examined for IL-10 mRNA accumulation by RT-PCR. Data represents mRNA samples from sorted cells isolated from pools of n=5 spleens and is shown as fold difference over naïve. One of three independent experiments is shown. B. Spleen cell suspensions from naïve BALB/c mice and BALB/c mice infected for 28d with L. donovani were stained for CD49b and NKp46. Plots are gated on CD49b+CD3- cells. C,D. Immunohistology of naïve and d28 infected (D) BALB/c spleen showing distribution of NKp46+ NK cells (green) and F4/80+ red pulp macrophages (red). Sections were counterstained with DAPI (blue). E. Hepatic NK cells co-express CD49b and NKp46. Cells from naïve and 28d-infected BALB/c mice were gated as CD49b+CD3-. For further characterisation, see Supplementary Figure 1. F. NKp46+ NK cells (green) co-localise with F4/80+ macrophages (blue) and CD3+ T cells (red) in hepatic granulomas caused by L. donovani. G,H. Frequency of granulomas identified as containing NKp46+ cells (G) and the frequency of NKp46+ cells located inside granulomas or outside granulomas i.e. in the parenchyma (H), was enumerated from approximately 35-40 granulomas per mouse (in 20 random 8μm sections). Data represents mean ±SD obtained from 3 mice.
) were sorted to >99% purity and then examined for IL-10 mRNA accumulation by RT-PCR. Data represents mRNA samples from sorted cells isolated from pools of n=5 spleens and is shown as fold difference over naïve. One of three independent experiments is shown. B. Spleen cell suspensions from naïve BALB/c mice and BALB/c mice infected for 28d with L. donovani were stained for CD49b and NKp46. Plots are gated on CD49b+CD3- cells. C,D. Immunohistology of naïve and d28 infected (D) BALB/c spleen showing distribution of NKp46+ NK cells (green) and F4/80+ red pulp macrophages (red). Sections were counterstained with DAPI (blue). E. Hepatic NK cells co-express CD49b and NKp46. Cells from naïve and 28d-infected BALB/c mice were gated as CD49b+CD3-. For further characterisation, see Supplementary Figure 1. F. NKp46+ NK cells (green) co-localise with F4/80+ macrophages (blue) and CD3+ T cells (red) in hepatic granulomas caused by L. donovani. G,H. Frequency of granulomas identified as containing NKp46+ cells (G) and the frequency of NKp46+ cells located inside granulomas or outside granulomas i.e. in the parenchyma (H), was enumerated from approximately 35-40 granulomas per mouse (in 20 random 8μm sections). Data represents mean ±SD obtained from 3 mice.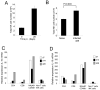
 ),14d (
),14d ( )or 28d (
)or 28d ( )with L. donovani. CD4+CD3+, CD8+CD3+ T cells, NKp46+CD49b+ and all residual cells (including DC, macrophages and stromal cells) were sorted to >99% purity and then examined for IL-10 mRNA accumulation by RT-PCR. Data represents mRNA samples from sorted cells isolated from pools of n=5 spleens and is shown as fold difference over naïve. Data from one of two independent experiments is shown.
)with L. donovani. CD4+CD3+, CD8+CD3+ T cells, NKp46+CD49b+ and all residual cells (including DC, macrophages and stromal cells) were sorted to >99% purity and then examined for IL-10 mRNA accumulation by RT-PCR. Data represents mRNA samples from sorted cells isolated from pools of n=5 spleens and is shown as fold difference over naïve. Data from one of two independent experiments is shown.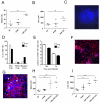


 ) . Supernatants were collected each day for 4 days and IL-10 (left panels) and IFNγ (right panels) was determined by ELISA. G. NK cells from mice infected for 24h (○) and 21d (□) were cultured in the presence of actinomycin D. At indicated times, IL-10 mRNA accumulation was determined. Data are expressed as % remaining IL-10 mRNA relative to mRNA levels at time 0 (168 vs 163 molecules of IL-10 mRNA / 1000 molecules HPRT in 24h and d21 samples respectively). Data represent one of two independent experiments.
) . Supernatants were collected each day for 4 days and IL-10 (left panels) and IFNγ (right panels) was determined by ELISA. G. NK cells from mice infected for 24h (○) and 21d (□) were cultured in the presence of actinomycin D. At indicated times, IL-10 mRNA accumulation was determined. Data are expressed as % remaining IL-10 mRNA relative to mRNA levels at time 0 (168 vs 163 molecules of IL-10 mRNA / 1000 molecules HPRT in 24h and d21 samples respectively). Data represent one of two independent experiments.Similar articles
-
An in vivo analysis of cytokine production during Leishmania donovani infection in scid mice.Exp Parasitol. 1996 Nov;84(2):195-202. doi: 10.1006/expr.1996.0105. Exp Parasitol. 1996. PMID: 8932769
-
Critical role of IRF-5 in the development of T helper 1 responses to Leishmania donovani infection.PLoS Pathog. 2011 Jan 6;7(1):e1001246. doi: 10.1371/journal.ppat.1001246. PLoS Pathog. 2011. PMID: 21253574 Free PMC article.
-
Granzyme-mediated regulation of host defense in the liver in experimental Leishmania donovani infection.Infect Immun. 2015 Feb;83(2):702-12. doi: 10.1128/IAI.02418-14. Epub 2014 Dec 1. Infect Immun. 2015. PMID: 25452549 Free PMC article.
-
Tissue granuloma structure-function in experimental visceral leishmaniasis.Int J Exp Pathol. 2001 Oct;82(5):249-67. doi: 10.1046/j.1365-2613.2001.00199.x. Int J Exp Pathol. 2001. PMID: 11703536 Free PMC article. Review.
-
Determinants of Innate Immunity in Visceral Leishmaniasis and Their Implication in Vaccine Development.Front Immunol. 2021 Oct 12;12:748325. doi: 10.3389/fimmu.2021.748325. eCollection 2021. Front Immunol. 2021. PMID: 34712235 Free PMC article. Review.
Cited by
-
T cell-derived IL-10 determines leishmaniasis disease outcome and is suppressed by a dendritic cell based vaccine.PLoS Pathog. 2013;9(6):e1003476. doi: 10.1371/journal.ppat.1003476. Epub 2013 Jun 27. PLoS Pathog. 2013. PMID: 23825956 Free PMC article.
-
Natural killer cells in experimental and human leishmaniasis.Front Cell Infect Microbiol. 2012 May 29;2:69. doi: 10.3389/fcimb.2012.00069. eCollection 2012. Front Cell Infect Microbiol. 2012. PMID: 22919660 Free PMC article. Review.
-
NK Cell IL-10 Production Requires IL-15 and IL-10 Driven STAT3 Activation.Front Immunol. 2019 Sep 4;10:2087. doi: 10.3389/fimmu.2019.02087. eCollection 2019. Front Immunol. 2019. PMID: 31552035 Free PMC article.
-
Regulation of mRNA stability contributes to the function of innate lymphoid cells in various diseases.Front Immunol. 2023 Jan 26;14:1118483. doi: 10.3389/fimmu.2023.1118483. eCollection 2023. Front Immunol. 2023. PMID: 36776864 Free PMC article. Review.
-
Leishmania-infected macrophages are targets of NK cell-derived cytokines but not of NK cell cytotoxicity.Infect Immun. 2011 Jul;79(7):2699-708. doi: 10.1128/IAI.00079-11. Epub 2011 Apr 25. Infect Immun. 2011. PMID: 21518784 Free PMC article.
References
-
- Alli RS, Khar A. Interleukin-12 secreted by mature dendritic cells mediates activation of NK cell function. FEBS Lett. 2004;559:71–76. - PubMed
-
- Andrews DM, Farrell HE, Densley EH, Scalzo AA, Shellam GR, Degli-Esposti MA. NK1.1+ cells and murine cytomegalovirus infection: what happens in situ? J Immunol. 2001;166:1796–1802. - PubMed
-
- Ato M, Stager S, Engwerda CR, Kaye PM. Defective CCR7 expression on dendritic cells contributes to the development of visceral leishmaniasis. Nat Immunol. 2002;3:1185–1191. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

