Catalytic mechanism of human DNA polymerase lambda with Mg2+ and Mn2+ from ab initio quantum mechanical/molecular mechanical studies
- PMID: 18692600
- PMCID: PMC2586819
- DOI: 10.1016/j.dnarep.2008.07.007
Catalytic mechanism of human DNA polymerase lambda with Mg2+ and Mn2+ from ab initio quantum mechanical/molecular mechanical studies
Abstract
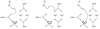
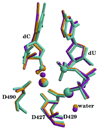
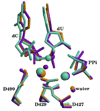
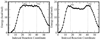
DNA polymerases play a crucial role in the cell cycle due to their involvement in genome replication and repair. Understanding the reaction mechanism by which these polymerases carry out their function can provide insights into these processes. Recently, the crystal structures of human DNA polymerase lambda (Pollambda) have been reported both for pre- and post-catalytic complexes [García-Díaz et al., DNA Repair 3 (2007), 1333]. Here we employ the pre-catalytic complex as a starting structure for the determination of the catalytic mechanism of Pollambda using ab initio quantum mechanical/molecular mechanical methods. The reaction path has been calculated using Mg(2+) and Mn(2+) as the catalytic metals. In both cases the reaction proceeds through a two-step mechanism where the 3'-OH of the primer sugar ring is deprotonated by one of the conserved Asp residues (D490) in the active site before the incorporation of the nucleotide to the nascent DNA chain. A significant charge transfer is observed between both metals and some residues in the active site as the reaction proceeds. The optimized reactant and product structures agree with the reported crystal structures. In addition, the calculated reaction barriers for both metals are close to experimentally estimated barriers. Energy decomposition analysis to explain individual residue contributions suggests that several amino acids surrounding the active site are important for catalysis. Some of these residues, including R420, R488 and E529, have been implicated in catalysis by previous mutagenesis experiments on the homologous residues on Polbeta. Furthermore, Pollambda residues R420 and E529 found to be important from the energy decomposition analysis, are homologous to residues R183 and E295 in Polbeta, both of which are linked to cancer. In addition, residues R386, E391, K422 and K472 appear to have an important role in catalysis and could be a potential target for mutagenesis experiments. There is partial conservation of these residues across the Pol X family of DNA polymerases.
Conflict of interest statement
Conflict of Interest statement
The authors declare that there are no conflicts of interest.
Figures
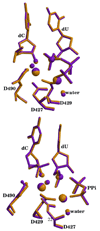


Similar articles
-
Role of the catalytic metal during polymerization by DNA polymerase lambda.DNA Repair (Amst). 2007 Sep 1;6(9):1333-40. doi: 10.1016/j.dnarep.2007.03.005. Epub 2007 May 1. DNA Repair (Amst). 2007. PMID: 17475573 Free PMC article.
-
A structural basis for metal ion mutagenicity and nucleotide selectivity in human DNA polymerase beta.Biochemistry. 1996 Oct 1;35(39):12762-77. doi: 10.1021/bi9529566. Biochemistry. 1996. PMID: 8841119
-
Structural basis for the inefficient nucleotide incorporation opposite cisplatin-DNA lesion by human DNA polymerase β.J Biol Chem. 2014 Nov 7;289(45):31341-8. doi: 10.1074/jbc.M114.605451. Epub 2014 Sep 18. J Biol Chem. 2014. PMID: 25237188 Free PMC article.
-
New structural snapshots provide molecular insights into the mechanism of high fidelity DNA synthesis.DNA Repair (Amst). 2015 Aug;32:3-9. doi: 10.1016/j.dnarep.2015.04.007. Epub 2015 Apr 30. DNA Repair (Amst). 2015. PMID: 26002198 Free PMC article. Review.
-
For the Better or for the Worse? The Effect of Manganese on the Activity of Eukaryotic DNA Polymerases.Int J Mol Sci. 2023 Dec 27;25(1):363. doi: 10.3390/ijms25010363. Int J Mol Sci. 2023. PMID: 38203535 Free PMC article. Review.
Cited by
-
Computational analysis of ammonia transfer along two intramolecular tunnels in Staphylococcus aureus glutamine-dependent amidotransferase (GatCAB).J Phys Chem B. 2015 Mar 5;119(9):3669-77. doi: 10.1021/jp5123568. Epub 2015 Feb 20. J Phys Chem B. 2015. PMID: 25654336 Free PMC article.
-
Relationship between conformational changes in pol lambda's active site upon binding incorrect nucleotides and mismatch incorporation rates.J Phys Chem B. 2009 Oct 1;113(39):13035-47. doi: 10.1021/jp903172x. J Phys Chem B. 2009. PMID: 19572669 Free PMC article.
-
Machines on Genes through the Computational Microscope.J Chem Theory Comput. 2023 Apr 11;19(7):1945-1964. doi: 10.1021/acs.jctc.2c01313. Epub 2023 Mar 22. J Chem Theory Comput. 2023. PMID: 36947696 Free PMC article. Review.
-
Reaction mechanism of the epsilon subunit of E. coli DNA polymerase III: insights into active site metal coordination and catalytically significant residues.J Am Chem Soc. 2009 Feb 4;131(4):1550-6. doi: 10.1021/ja8082818. J Am Chem Soc. 2009. PMID: 19119875 Free PMC article.
-
Preferred WMSA catalytic mechanism of the nucleotidyl transfer reaction in human DNA polymerase κ elucidates error-free bypass of a bulky DNA lesion.Nucleic Acids Res. 2012 Oct;40(18):9193-205. doi: 10.1093/nar/gks653. Epub 2012 Jul 5. Nucleic Acids Res. 2012. PMID: 22772988 Free PMC article.
References
-
- Iwanaga A, Ouchida M, Miyazaki K, Hori K, Mukai T. Functional mutation of DNA polymerase β found in human gastric cancer - inability of the base excision repair in vivo. Mutation Res. 1999;vol. 435:121–128. - PubMed
-
- Starcevic D, Dalal S, Sweasy JB. Is there a link between DNA polymerase β and cancer? Cell Cycle. 2004;vol. 3:98–1001. - PubMed
-
- Kunkel TA. Considering the cancer consequences of altered DNA polymerase function. Cancer Cell. 2003;vol. 3:105–110. - PubMed
-
- Bebenek K, Kunkel TA. Functions of DNA polymerases. Adv. In Prot. Chem. 2004;vol. 69:137–165. - PubMed
-
- Cowan J. Structural and catalytic chemistry of magnesium-dependent enzymes. BioMetals. 2002;vol. 15:225–235. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

