Functional divergence between co-chaperones of Hsc70
- PMID: 18684711
- PMCID: PMC5026489
- DOI: 10.1074/jbc.M803923200
Functional divergence between co-chaperones of Hsc70
Abstract
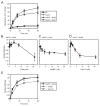
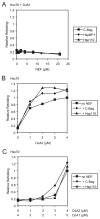
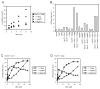
The ATPase cycle of the chaperone Hsc70 is regulated by co-chaperones; Hsp40/DnaJ-related proteins stimulate ATP hydrolysis by Hsc70 and can bind unfolded polypeptides themselves. Conversely, various nucleotide exchange factors (NEFs) stimulate ADP-ATP exchange by Hsc70. We analyzed the purified Hsp40-related co-chaperones DJA1 (Hdj2) and DJA2 (Hdj3) and found that they had a distinct pattern of binding to a range of polypeptides. DJA2 alone could stimulate Hsc70-mediated refolding of luciferase in the absence of NEF, whereas DJA1 was much less active. The addition of the Bag1 NEF increased refolding by Hsc70 and DJA2, as did the newly characterized NEF Hsp110, but each NEF had a different optimal concentration ratio to Hsc70. Notably, the NEF HspBP1 could not increase refolding by Hsc70 and DJA2 at any concentration, and none of the NEFs improved the refolding activity with DJA1. Instead, DJA1 was inhibitory of refolding with DJA2 and Hsc70. All combinations of DJA1 or DJA2 with the three NEFs stimulated the Hsc70 ATPase rate, although Hsp110 became less effective with increasing concentrations. A chimeric DJA2 having its Hsc70-stimulatory J domain replaced with that of DJA1 was functional for polypeptide binding and ATPase stimulation of Hsc70. However, it could not support efficient Hsc70-mediated refolding and also inhibited refolding with DJA2 and Hsc70. These results suggest a more complex model of Hsc70 mechanism than has been previously thought, with notable functional divergence between Hsc70 co-chaperones.
Figures
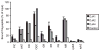
Similar articles
-
The DNAJA2 substrate release mechanism is essential for chaperone-mediated folding.J Biol Chem. 2012 Dec 7;287(50):41939-54. doi: 10.1074/jbc.M112.413278. Epub 2012 Oct 22. J Biol Chem. 2012. PMID: 23091061 Free PMC article.
-
Binding of human nucleotide exchange factors to heat shock protein 70 (Hsp70) generates functionally distinct complexes in vitro.J Biol Chem. 2014 Jan 17;289(3):1402-14. doi: 10.1074/jbc.M113.521997. Epub 2013 Dec 5. J Biol Chem. 2014. PMID: 24318877 Free PMC article.
-
Multiple molecules of Hsc70 and a dimer of DjA1 independently bind to an unfolded protein.J Biol Chem. 2010 May 28;285(22):16789-97. doi: 10.1074/jbc.M110.101501. Epub 2010 Apr 2. J Biol Chem. 2010. PMID: 20363747 Free PMC article.
-
Nucleotide Exchange Factors for Hsp70 Molecular Chaperones: GrpE, Hsp110/Grp170, HspBP1/Sil1, and BAG Domain Proteins.Subcell Biochem. 2023;101:1-39. doi: 10.1007/978-3-031-14740-1_1. Subcell Biochem. 2023. PMID: 36520302 Review.
-
GrpE, Hsp110/Grp170, HspBP1/Sil1 and BAG domain proteins: nucleotide exchange factors for Hsp70 molecular chaperones.Subcell Biochem. 2015;78:1-33. doi: 10.1007/978-3-319-11731-7_1. Subcell Biochem. 2015. PMID: 25487014 Review.
Cited by
-
hERG quality control and the long QT syndrome.J Physiol. 2016 May 1;594(9):2469-81. doi: 10.1113/JP270531. Epub 2016 Feb 9. J Physiol. 2016. PMID: 26718903 Free PMC article. Review.
-
The role of molecular chaperones in clathrin mediated vesicular trafficking.Front Mol Biosci. 2015 May 19;2:26. doi: 10.3389/fmolb.2015.00026. eCollection 2015. Front Mol Biosci. 2015. PMID: 26042225 Free PMC article. Review.
-
Stress-induced p53 drives BAG5 cochaperone expression to control α-synuclein aggregation in Parkinson's disease.Aging (Albany NY). 2020 Oct 21;12(20):20702-20727. doi: 10.18632/aging.103998. Epub 2020 Oct 21. Aging (Albany NY). 2020. PMID: 33085644 Free PMC article.
-
Hsp70 and DNAJA2 limit CFTR levels through degradation.PLoS One. 2019 Aug 13;14(8):e0220984. doi: 10.1371/journal.pone.0220984. eCollection 2019. PLoS One. 2019. PMID: 31408507 Free PMC article.
-
Crucial HSP70 co-chaperone complex unlocks metazoan protein disaggregation.Nature. 2015 Aug 13;524(7564):247-51. doi: 10.1038/nature14884. Epub 2015 Aug 5. Nature. 2015. PMID: 26245380 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

