Genomic analysis of the TRIM family reveals two groups of genes with distinct evolutionary properties
- PMID: 18673550
- PMCID: PMC2533329
- DOI: 10.1186/1471-2148-8-225
Genomic analysis of the TRIM family reveals two groups of genes with distinct evolutionary properties
Abstract
Background: The TRIM family is composed of multi-domain proteins that display the Tripartite Motif (RING, B-box and Coiled-coil) that can be associated with a C-terminal domain. TRIM genes are involved in ubiquitylation and are implicated in a variety of human pathologies, from Mendelian inherited disorders to cancer, and are also involved in cellular response to viral infection.
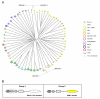
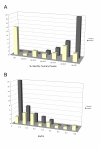
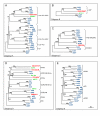
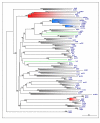
Results: Here we defined the entire human TRIM family and also identified the TRIM sets of other vertebrate (mouse, rat, dog, cow, chicken, tetraodon, and zebrafish) and invertebrate species (fruitfly, worm, and ciona). By means of comparative analyses we found that, after assembly of the tripartite motif in an early metazoan ancestor, few types of C-terminal domains have been associated with this module during evolution and that an important increase in TRIM number occurred in vertebrate species concomitantly with the addition of the SPRY domain. We showed that the human TRIM family is split into two groups that differ in domain structure, genomic organization and evolutionary properties. Group 1 members present a variety of C-terminal domains, are highly conserved among vertebrate species, and are represented in invertebrates. Conversely, group 2 is absent in invertebrates, is characterized by the presence of a C-terminal SPRY domain and presents unique sets of genes in each mammal examined. The generation of independent sets of group 2 genes is also evident in the other vertebrate species. Comparing the murine and human TRIM sets, we found that group 1 and 2 genes evolve at different speeds and are subject to different selective pressures.
Conclusion: We found that the TRIM family is composed of two groups of genes with distinct evolutionary properties. Group 2 is younger, highly dynamic, and might act as a reservoir to develop novel TRIM functions. Since some group 2 genes are implicated in innate immune response, their evolutionary features may account for species-specific battles against viral infection.
Figures


Similar articles
-
Genomics and evolution of the TRIM gene family.Adv Exp Med Biol. 2012;770:1-9. doi: 10.1007/978-1-4614-5398-7_1. Adv Exp Med Biol. 2012. PMID: 23630996 Review.
-
Genomic Analysis, Evolution and Characterization of E3 Ubiquitin Protein Ligase (TRIM) Gene Family in Common Carp (Cyprinus carpio).Genes (Basel). 2023 Mar 7;14(3):667. doi: 10.3390/genes14030667. Genes (Basel). 2023. PMID: 36980939 Free PMC article.
-
Origin and evolution of TRIM proteins: new insights from the complete TRIM repertoire of zebrafish and pufferfish.PLoS One. 2011;6(7):e22022. doi: 10.1371/journal.pone.0022022. Epub 2011 Jul 15. PLoS One. 2011. PMID: 21789205 Free PMC article.
-
Evolution of a TRIM5-CypA splice isoform in old world monkeys.PLoS Pathog. 2008 Feb 29;4(2):e1000003. doi: 10.1371/journal.ppat.1000003. PLoS Pathog. 2008. PMID: 18389077 Free PMC article.
-
TRIM proteins as RING finger E3 ubiquitin ligases.Adv Exp Med Biol. 2012;770:27-37. doi: 10.1007/978-1-4614-5398-7_3. Adv Exp Med Biol. 2012. PMID: 23630998 Review.
Cited by
-
TRIM Proteins in Host Defense and Viral Pathogenesis.Curr Clin Microbiol Rep. 2020;7(4):101-114. doi: 10.1007/s40588-020-00150-8. Epub 2020 Aug 8. Curr Clin Microbiol Rep. 2020. PMID: 32837832 Free PMC article. Review.
-
Evolution and expression of the duck TRIM gene repertoire.Front Immunol. 2023 Aug 9;14:1220081. doi: 10.3389/fimmu.2023.1220081. eCollection 2023. Front Immunol. 2023. PMID: 37622121 Free PMC article.
-
Regulation of innate immune signalling pathways by the tripartite motif (TRIM) family proteins.EMBO Mol Med. 2011 Sep;3(9):513-27. doi: 10.1002/emmm.201100160. Epub 2011 Aug 9. EMBO Mol Med. 2011. PMID: 21826793 Free PMC article. Review.
-
Loss of TRIM62 expression is an independent adverse prognostic factor in acute myeloid leukemia.Clin Lymphoma Myeloma Leuk. 2015 Feb;15(2):115-127.e15. doi: 10.1016/j.clml.2014.07.011. Epub 2014 Aug 12. Clin Lymphoma Myeloma Leuk. 2015. PMID: 25248926 Free PMC article.
-
Genome editing of FTR42 improves zebrafish survival against virus infection by enhancing IFN immunity.iScience. 2024 Mar 12;27(4):109497. doi: 10.1016/j.isci.2024.109497. eCollection 2024 Apr 19. iScience. 2024. PMID: 38550983 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

