CTCF regulates allelic expression of Igf2 by orchestrating a promoter-polycomb repressive complex 2 intrachromosomal loop
- PMID: 18662993
- PMCID: PMC2577414
- DOI: 10.1128/MCB.00204-08
CTCF regulates allelic expression of Igf2 by orchestrating a promoter-polycomb repressive complex 2 intrachromosomal loop
Abstract
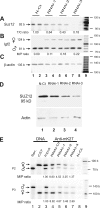
CTCF is a zinc finger DNA-binding protein that regulates the epigenetic states of numerous target genes. Using allelic regulation of mouse insulin-like growth factor II (Igf2) as a model, we demonstrate that CTCF binds to the unmethylated maternal allele of the imprinting control region (ICR) in the Igf2/H19 imprinting domain and forms a long-range intrachromosomal loop to interact with the three clustered Igf2 promoters. Polycomb repressive complex 2 is recruited through the interaction of CTCF with Suz12, leading to allele-specific methylation at lysine 27 of histone H3 (H3-K27) and to suppression of the maternal Igf2 promoters. Targeted mutation or deletion of the maternal ICR abolishes this chromatin loop, decreases allelic H3-K27 methylation, and causes loss of Igf2 imprinting. RNA interference knockdown of Suz12 also leads to reactivation of the maternal Igf2 allele and biallelic Igf2 expression. CTCF and Suz12 are coprecipitated from nuclear extracts with antibodies specific for either protein, and they interact with each other in a two-hybrid system. These findings offer insight into general epigenetic mechanisms by which CTCF governs gene expression by orchestrating chromatin loop structures and by serving as a DNA-binding protein scaffold to recruit and bind polycomb repressive complexes.
Figures




Similar articles
-
Interruption of intrachromosomal looping by CCCTC binding factor decoy proteins abrogates genomic imprinting of human insulin-like growth factor II.J Cell Biol. 2011 May 2;193(3):475-87. doi: 10.1083/jcb.201101021. J Cell Biol. 2011. PMID: 21536749 Free PMC article.
-
Promoter histone H3K27 methylation in the control of IGF2 imprinting in human tumor cell lines.Hum Mol Genet. 2014 Jan 1;23(1):117-28. doi: 10.1093/hmg/ddt405. Epub 2013 Aug 19. Hum Mol Genet. 2014. PMID: 23962719 Free PMC article.
-
Restoration of IGF2 imprinting by polycomb repressive complex 2 docking factor SUZ12 in colon cancer cells.Exp Cell Res. 2015 Nov 1;338(2):214-21. doi: 10.1016/j.yexcr.2015.09.016. Epub 2015 Sep 25. Exp Cell Res. 2015. PMID: 26407907
-
Genomic imprinting: CTCF protects the boundaries.Curr Biol. 2004 Apr 6;14(7):R284-6. doi: 10.1016/j.cub.2004.03.026. Curr Biol. 2004. PMID: 15062124 Review.
-
Transcriptional control: imprinting insulation.Curr Biol. 2000 Jun 15;10(12):R463-5. doi: 10.1016/s0960-9822(00)00534-0. Curr Biol. 2000. PMID: 10873799 Review.
Cited by
-
CTCF-mediated chromatin looping in EGR2 regulation and SUZ12 recruitment critical for peripheral myelination and repair.Nat Commun. 2020 Aug 17;11(1):4133. doi: 10.1038/s41467-020-17955-2. Nat Commun. 2020. PMID: 32807777 Free PMC article.
-
Gene signatures in hepatocellular carcinoma (HCC).Semin Cancer Biol. 2011 Feb;21(1):4-9. doi: 10.1016/j.semcancer.2010.09.002. Epub 2010 Sep 17. Semin Cancer Biol. 2011. PMID: 20851183 Free PMC article. Review.
-
Chromatin structure and the inheritance of epigenetic information.Nat Rev Genet. 2010 Apr;11(4):285-96. doi: 10.1038/nrg2752. Nat Rev Genet. 2010. PMID: 20300089 Free PMC article. Review.
-
Allele-specific H3K79 Di- versus trimethylation distinguishes opposite parental alleles at imprinted regions.Mol Cell Biol. 2010 Jun;30(11):2693-707. doi: 10.1128/MCB.01537-09. Epub 2010 Mar 29. Mol Cell Biol. 2010. PMID: 20351169 Free PMC article.
-
Shaping the Genome with Non-Coding RNAs.Curr Genomics. 2011 Aug;12(5):307-21. doi: 10.2174/138920211796429772. Curr Genomics. 2011. PMID: 21874119 Free PMC article.
References
-
- Arney, K. L. 2003. H19 and Igf2—enhancing the confusion? Trends Genet. 1917-23. - PubMed
-
- Bartolomei, M. S. 2003. Epigenetics: role of germ cell imprinting. Adv. Exp. Med. Biol. 518239-245. - PubMed
-
- Bartolomei, M. S., A. L. Webber, M. E. Brunkow, and S. M. Tilghman. 1993. Epigenetic mechanisms underlying the imprinting of the mouse H19 gene. Genes Dev. 71663-1673. - PubMed
-
- Bell, A. C., and G. Felsenfeld. 2000. Methylation of a CTCF-dependent boundary controls imprinted expression of the Igf2 gene. Nature 405482-485. - PubMed
-
- Bell, A. C., A. G. West, and G. Felsenfeld. 2001. Insulators and boundaries: versatile regulatory elements in the eukaryotic genome. Science 291447-450. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous
