Mistranslation-induced protein misfolding as a dominant constraint on coding-sequence evolution
- PMID: 18662548
- PMCID: PMC2696314
- DOI: 10.1016/j.cell.2008.05.042
Mistranslation-induced protein misfolding as a dominant constraint on coding-sequence evolution
Abstract
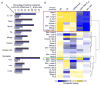
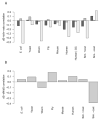
Strikingly consistent correlations between rates of coding-sequence evolution and gene expression levels are apparent across taxa, but the biological causes behind the selective pressures on coding-sequence evolution remain controversial. Here, we demonstrate conserved patterns of simple covariation between sequence evolution, codon usage, and mRNA level in E. coli, yeast, worm, fly, mouse, and human that suggest that all observed trends stem largely from a unified underlying selective pressure. In metazoans, these trends are strongest in tissues composed of neurons, whose structure and lifetime confer extreme sensitivity to protein misfolding. We propose, and demonstrate using a molecular-level evolutionary simulation, that selection against toxicity of misfolded proteins generated by ribosome errors suffices to create all of the observed covariation. The mechanistic model of molecular evolution that emerges yields testable biochemical predictions, calls into question the use of nonsynonymous-to-synonymous substitution ratios (Ka/Ks) to detect functional selection, and suggests how mistranslation may contribute to neurodegenerative disease.
Figures

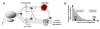


Similar articles
-
Translationally optimal codons associate with structurally sensitive sites in proteins.Mol Biol Evol. 2009 Jul;26(7):1571-80. doi: 10.1093/molbev/msp070. Epub 2009 Apr 6. Mol Biol Evol. 2009. PMID: 19349643 Free PMC article.
-
Secreted Proteins Defy the Expression Level-Evolutionary Rate Anticorrelation.Mol Biol Evol. 2017 Mar 1;34(3):692-706. doi: 10.1093/molbev/msw268. Mol Biol Evol. 2017. PMID: 28007979 Free PMC article.
-
Natural selection against protein aggregation on self-interacting and essential proteins in yeast, fly, and worm.Mol Biol Evol. 2008 Aug;25(8):1530-3. doi: 10.1093/molbev/msn122. Epub 2008 May 23. Mol Biol Evol. 2008. PMID: 18503047 Free PMC article.
-
Evolution of synonymous codon usage in metazoans.Curr Opin Genet Dev. 2002 Dec;12(6):640-9. doi: 10.1016/s0959-437x(02)00353-2. Curr Opin Genet Dev. 2002. PMID: 12433576 Review.
-
Neurodegenerative diseases: Lessons from genome-wide screens in small model organisms.EMBO Mol Med. 2009 Nov;1(8-9):360-70. doi: 10.1002/emmm.200900051. EMBO Mol Med. 2009. PMID: 20049741 Free PMC article. Review.
Cited by
-
The ribosome as a hub for protein quality control.Mol Cell. 2013 Feb 7;49(3):411-21. doi: 10.1016/j.molcel.2013.01.020. Mol Cell. 2013. PMID: 23395271 Free PMC article. Review.
-
Improved prediction of site-rates from structure with averaging across homologs.Protein Sci. 2024 Jul;33(7):e5086. doi: 10.1002/pro.5086. Protein Sci. 2024. PMID: 38923241 Free PMC article.
-
Impact of Synonymous Genome Recoding on the HIV Life Cycle.Front Microbiol. 2021 Mar 16;12:606087. doi: 10.3389/fmicb.2021.606087. eCollection 2021. Front Microbiol. 2021. PMID: 33796084 Free PMC article. Review.
-
Codon Usage Bias: A Potential Factor Affecting VGLUT Developmental Expression and Protein Evolution.Mol Neurobiol. 2024 Sep 21. doi: 10.1007/s12035-024-04426-8. Online ahead of print. Mol Neurobiol. 2024. PMID: 39305444
-
Cancer, Warts, or Asymptomatic Infections: Clinical Presentation Matches Codon Usage Preferences in Human Papillomaviruses.Genome Biol Evol. 2015 Jul 1;7(8):2117-35. doi: 10.1093/gbe/evv129. Genome Biol Evol. 2015. PMID: 26139833 Free PMC article.
References
-
- Anfinsen C. The Molecular Basis of Evolution. New York: John Wiley & Sons, Inc; 1959.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

