Prion protein amino acid determinants of differential susceptibility and molecular feature of prion strains in mice and voles
- PMID: 18654630
- PMCID: PMC2453331
- DOI: 10.1371/journal.ppat.1000113
Prion protein amino acid determinants of differential susceptibility and molecular feature of prion strains in mice and voles
Abstract
The bank vole is a rodent susceptible to different prion strains from humans and various animal species. We analyzed the transmission features of different prions in a panel of seven rodent species which showed various degrees of phylogenetic affinity and specific prion protein (PrP) sequence divergences in order to investigate the basis of vole susceptibility in comparison to other rodent models. At first, we found a differential susceptibility of bank and field voles compared to C57Bl/6 and wood mice. Voles showed high susceptibility to sheep scrapie but were resistant to bovine spongiform encephalopathy, whereas C57Bl/6 and wood mice displayed opposite features. Infection with mouse-adapted scrapie 139A was faster in voles than in C57Bl/6 and wood mice. Moreover, a glycoprofile change was observed in voles, which was reverted upon back passage to mice. All strains replicated much faster in voles than in mice after adapting to the new species. PrP sequence comparison indicated a correlation between the transmission patterns and amino acids at positions 154 and 169 (Y and S in mice, N and N in voles). This correlation was confirmed when inoculating three additional rodent species: gerbils, spiny mice and oldfield mice with sheep scrapie and 139A. These rodents were chosen because oldfield mice do have the 154N and 169N substitutions, whereas gerbil and spiny mice do not have them. Our results suggest that PrP residues 154 and 169 drive the susceptibility, molecular phenotype and replication rate of prion strains in rodents. This might have implications for the assessment of host range and molecular traceability of prion strains, as well as for the development of improved animal models for prion diseases.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures
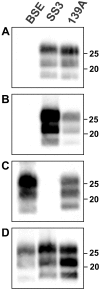
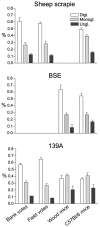






Similar articles
-
PrPC Governs Susceptibility to Prion Strains in Bank Vole, While Other Host Factors Modulate Strain Features.J Virol. 2016 Nov 14;90(23):10660-10669. doi: 10.1128/JVI.01592-16. Print 2016 Dec 1. J Virol. 2016. PMID: 27654300 Free PMC article.
-
Conversion efficiency of bank vole prion protein in vitro is determined by residues 155 and 170, but does not correlate with the high susceptibility of bank voles to sheep scrapie in vivo.J Biol Chem. 2006 Apr 7;281(14):9373-84. doi: 10.1074/jbc.M512239200. Epub 2006 Feb 2. J Biol Chem. 2006. PMID: 16455657
-
The bank vole (Myodes glareolus) as a sensitive bioassay for sheep scrapie.J Gen Virol. 2008 Dec;89(Pt 12):2975-2985. doi: 10.1099/vir.0.2008/005520-0. J Gen Virol. 2008. PMID: 19008382
-
Genetic and infectious prion diseases.Arch Neurol. 1993 Nov;50(11):1129-53. doi: 10.1001/archneur.1993.00540110011002. Arch Neurol. 1993. PMID: 8105771 Review.
-
Prion encephalopathies of animals and humans.Dev Biol Stand. 1993;80:31-44. Dev Biol Stand. 1993. PMID: 8270114 Review.
Cited by
-
PrPC Governs Susceptibility to Prion Strains in Bank Vole, While Other Host Factors Modulate Strain Features.J Virol. 2016 Nov 14;90(23):10660-10669. doi: 10.1128/JVI.01592-16. Print 2016 Dec 1. J Virol. 2016. PMID: 27654300 Free PMC article.
-
Proteinase K-resistant material in ARR/VRQ sheep brain affected with classical scrapie is composed mainly of VRQ prion protein.J Virol. 2011 Dec;85(23):12537-46. doi: 10.1128/JVI.00448-11. Epub 2011 Sep 14. J Virol. 2011. PMID: 21917981 Free PMC article.
-
Further characterisation of transmissible spongiform encephalopathy phenotypes after inoculation of cattle with two temporally separated sources of sheep scrapie from Great Britain.BMC Res Notes. 2015 Jul 24;8:312. doi: 10.1186/s13104-015-1260-3. BMC Res Notes. 2015. PMID: 26205536 Free PMC article.
-
Convergent generation of atypical prions in knockin mouse models of genetic prion disease.J Clin Invest. 2024 Aug 1;134(15):e176344. doi: 10.1172/JCI176344. J Clin Invest. 2024. PMID: 39087478 Free PMC article.
-
Variable Protease-Sensitive Prionopathy Transmission to Bank Voles.Emerg Infect Dis. 2019 Jan;25(1):73-81. doi: 10.3201/eid2501.180807. Emerg Infect Dis. 2019. PMID: 30561322 Free PMC article.
References
-
- Oesch B, Westaway D, Walchli M, McKinley MP, Kent SB, et al. A cellular gene encodes PrP 27–30 protein. Cell. 1985;40:735–746. - PubMed
-
- Prusiner SB. Molecular biology of prion diseases. Science. 1991;252:1515–1522. - PubMed
-
- Collinge J. Prion diseases of humans and animals: their causes and molecular basis. Annu Rev Neurosci. 2001;24:519–550. - PubMed
-
- Prusiner SB, Scott M, Foster D, Pan KM, Groth D, et al. Transgenetic studies implicate interactions between homologous PrP isoforms in scrapie prion replication. Cell. 1990;63:673–686. - PubMed
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Research Materials

