Stability and shape of hepatitis B virus capsids in vacuo
- PMID: 18642251
- PMCID: PMC2750006
- DOI: 10.1002/anie.200802410
Stability and shape of hepatitis B virus capsids in vacuo
Figures
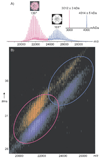
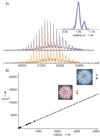
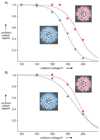
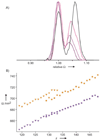
Similar articles
-
High plasticity of the hepatitis B virus capsid revealed by conformational stress.J Mol Biol. 2006 Feb 24;356(3):812-22. doi: 10.1016/j.jmb.2005.11.053. Epub 2005 Dec 5. J Mol Biol. 2006. PMID: 16378623
-
Kinetics versus Thermodynamics in Virus Capsid Polymorphism.J Phys Chem B. 2016 Jul 7;120(26):6003-9. doi: 10.1021/acs.jpcb.6b01953. Epub 2016 Apr 8. J Phys Chem B. 2016. PMID: 27027925
-
Interrogating viral capsid assembly with ion mobility-mass spectrometry.Nat Chem. 2011 Feb;3(2):126-32. doi: 10.1038/nchem.947. Epub 2010 Dec 19. Nat Chem. 2011. PMID: 21258385
-
Hepatitis B Core Protein Capsids.Subcell Biochem. 2021;96:451-470. doi: 10.1007/978-3-030-58971-4_14. Subcell Biochem. 2021. PMID: 33252740 Review.
-
Bacteriophage HK97 capsid assembly and maturation.Adv Exp Med Biol. 2012;726:351-63. doi: 10.1007/978-1-4614-0980-9_15. Adv Exp Med Biol. 2012. PMID: 22297521 Review.
Cited by
-
Are the Gas-Phase Structures of Molecular Elephants Enduring or Ephemeral? Results from Time-Dependent, Tandem Ion Mobility.Anal Chem. 2023 Jun 27;95(25):9589-9597. doi: 10.1021/acs.analchem.3c01222. Epub 2023 Jun 9. Anal Chem. 2023. PMID: 37294019 Free PMC article.
-
Ion Mobility Spectrometry-Mass Spectrometry of Intrinsically Unfolded Proteins: Trying to Put Order into Disorder.Curr Anal Chem. 2013 Apr;9(2):181-191. doi: 10.2174/1573411011309020004. Curr Anal Chem. 2013. PMID: 23885220 Free PMC article.
-
How hot are your ions in TWAVE ion mobility spectrometry?J Am Soc Mass Spectrom. 2012 Mar;23(3):553-62. doi: 10.1007/s13361-011-0313-7. Epub 2011 Dec 28. J Am Soc Mass Spectrom. 2012. PMID: 22203576 Free PMC article.
-
Subunit exchange rates in Hepatitis B virus capsids are geometry- and temperature-dependent.Phys Chem Chem Phys. 2010 Nov 7;12(41):13368-71. doi: 10.1039/c0cp00692k. Epub 2010 Jul 30. Phys Chem Chem Phys. 2010. PMID: 20676421 Free PMC article.
-
Determining Energies and Cross Sections of Individual Ions Using Higher-Order Harmonics in Fourier Transform Charge Detection Mass Spectrometry (FT-CDMS).J Am Soc Mass Spectrom. 2018 Sep;29(9):1861-1869. doi: 10.1007/s13361-018-1987-x. Epub 2018 Jun 2. J Am Soc Mass Spectrom. 2018. PMID: 29860679
References
-
- Blumberg BS. Proc. Natl. Acad. Sci. USA. 1997;94:7121. - DOI - PMC - PubMed
-
- Singh P, Gonzalez MJ, Manchester M. Drug Dev. Res. 2006;67:23. - DOI
-
- Caspar DL, Klug A. Cold Spring Harbor Symp. Quant. Biol. 1962;27:1. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

