Ribosomal mutations cause p53-mediated dark skin and pleiotropic effects
- PMID: 18641651
- PMCID: PMC3979291
- DOI: 10.1038/ng.188
Ribosomal mutations cause p53-mediated dark skin and pleiotropic effects
Abstract
Mutations in genes encoding ribosomal proteins cause the Minute phenotype in Drosophila and mice, and Diamond-Blackfan syndrome in humans. Here we report two mouse dark skin (Dsk) loci caused by mutations in Rps19 (ribosomal protein S19) and Rps20 (ribosomal protein S20). We identify a common pathophysiologic program in which p53 stabilization stimulates Kit ligand expression, and, consequently, epidermal melanocytosis via a paracrine mechanism. Accumulation of p53 also causes reduced body size and erythrocyte count. These results provide a mechanistic explanation for the diverse collection of phenotypes that accompany reduced dosage of genes encoding ribosomal proteins, and have implications for understanding normal human variation and human disease.
Figures


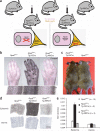

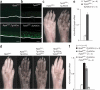
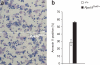
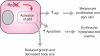
Comment in
-
Dark skin mutations shed light on inherited anemia.Nat Genet. 2008 Aug;40(8):931-2. doi: 10.1038/ng0808-931. Nat Genet. 2008. PMID: 18665127 No abstract available.
Similar articles
-
The essential role of p53 in hyperpigmentation of the skin via regulation of paracrine melanogenic cytokine receptor signaling.J Biol Chem. 2009 Feb 13;284(7):4343-53. doi: 10.1074/jbc.M805570200. Epub 2008 Dec 18. J Biol Chem. 2009. PMID: 19098008
-
SPRY1 Deficiency in Keratinocytes Induces Follicular Melanocyte Stem Cell Migration to the Epidermis through p53/Stem Cell Factor/C-KIT Signaling.J Invest Dermatol. 2024 Oct;144(10):2255-2266.e4. doi: 10.1016/j.jid.2024.02.018. Epub 2024 Mar 8. J Invest Dermatol. 2024. PMID: 38462125
-
CK1α ablation in keratinocytes induces p53-dependent, sunburn-protective skin hyperpigmentation.Proc Natl Acad Sci U S A. 2017 Sep 19;114(38):E8035-E8044. doi: 10.1073/pnas.1702763114. Epub 2017 Sep 6. Proc Natl Acad Sci U S A. 2017. PMID: 28878021 Free PMC article.
-
Nucleolar stress in Diamond Blackfan anemia pathophysiology.Biochim Biophys Acta. 2014 Jun;1842(6):765-8. doi: 10.1016/j.bbadis.2013.12.013. Epub 2014 Jan 8. Biochim Biophys Acta. 2014. PMID: 24412987 Review.
-
Haploinsufficiency of ribosomal proteins and p53 activation in anemia: Diamond-Blackfan anemia and the 5q- syndrome.Adv Biol Regul. 2012 Jan;52(1):196-203. doi: 10.1016/j.advenzreg.2011.09.008. Adv Biol Regul. 2012. PMID: 21930148 Review. No abstract available.
Cited by
-
Hematopoietic defects in rps29 mutant zebrafish depend upon p53 activation.Exp Hematol. 2012 Mar;40(3):228-237.e5. doi: 10.1016/j.exphem.2011.11.007. Epub 2011 Nov 25. Exp Hematol. 2012. PMID: 22120640 Free PMC article.
-
Important genes in the pathogenesis of 5q- syndrome and their connection with ribosomal stress and the innate immune system pathway.Leuk Res Treatment. 2012;2012:179402. doi: 10.1155/2012/179402. Epub 2012 Feb 13. Leuk Res Treatment. 2012. PMID: 23213547 Free PMC article.
-
RPSA, a candidate gene for isolated congenital asplenia, is required for pre-rRNA processing and spleen formation in Xenopus.Development. 2018 Oct 18;145(20):dev166181. doi: 10.1242/dev.166181. Development. 2018. PMID: 30337486 Free PMC article.
-
The p53 tumor suppressor protein is a critical regulator of hematopoietic stem cell behavior.Cell Cycle. 2009 Oct 1;8(19):3120-4. doi: 10.4161/cc.8.19.9627. Cell Cycle. 2009. PMID: 19755852 Free PMC article. Review.
-
Changes in the Transcriptome Caused by Mutations in the Ribosomal Protein uS10 Associated with a Predisposition to Colorectal Cancer.Int J Mol Sci. 2022 May 31;23(11):6174. doi: 10.3390/ijms23116174. Int J Mol Sci. 2022. PMID: 35682850 Free PMC article.
References
-
- Lambertsson A. The minute genes in Drosophila and their molecular functions. Adv. Genet. 1998;38:69–134. - PubMed
-
- Draptchinskaia N, et al. The gene encoding ribosomal protein S19 is mutated in Diamond-Blackfan anaemia. Nat. Genet. 1999;21:169–175. - PubMed
-
- Cmejla R, Cmejlova J, Handrkova H, Petrak J, Pospisilova D. Ribosomal protein S17 gene (RPS17) is mutated in Diamond-Blackfan anemia. Hum. Mutat. 2007;28:1178–1182. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

