CTCFL/BORIS is a methylation-independent DNA-binding protein that preferentially binds to the paternal H19 differentially methylated region
- PMID: 18632606
- PMCID: PMC2731476
- DOI: 10.1158/0008-5472.CAN-08-1005
CTCFL/BORIS is a methylation-independent DNA-binding protein that preferentially binds to the paternal H19 differentially methylated region
Abstract
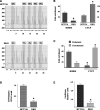
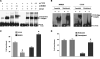
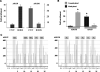
The CTCF paralog BORIS (brother of the regulator of imprinted sites) is an insulator DNA-binding protein thought to play a role in chromatin organization and gene expression. Under normal physiologic conditions, BORIS is predominantly expressed during embryonic male germ cell development; however, it is also expressed in tumors and tumor cell lines and, as such, has been classified as a cancer-germline or cancer-testis gene. It has been suggested that BORIS may be a pro-proliferative factor, whereas CTCF favors antiproliferation. BORIS and CTCF share similar zinc finger DNA-binding domains and seem to bind to identical target sequences. Thus, one critical question is the mechanism governing the DNA-binding specificity of these two proteins when both are present in tumor cells. Chromatin immunoprecipitation (ChIP) in HCT116 cells and their hypermethylated variant showed that BORIS binds to methylated DNA sequences, whereas CTCF binds to unmethylated DNA. Electromobility shift assays, using both whole-cell extracts and in vitro translated CTCF and BORIS protein, and methylation-specific ChIP PCR showed that BORIS is a methylation-independent DNA-binding protein. Finally, experiments in murine hybrid cells containing either the maternal or paternal human chromosome 11 showed that BORIS preferentially binds to the methylated paternal H19 differentially methylated region, suggesting a mechanism in which the affinity of CTCF for the unmethylated maternal allele directs the DNA binding of BORIS toward the paternal allele.
Figures
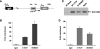
Similar articles
-
The testis-specific factor CTCFL cooperates with the protein methyltransferase PRMT7 in H19 imprinting control region methylation.PLoS Biol. 2006 Oct;4(11):e355. doi: 10.1371/journal.pbio.0040355. PLoS Biol. 2006. PMID: 17048991 Free PMC article.
-
Conditional expression of the CTCF-paralogous transcriptional factor BORIS in normal cells results in demethylation and derepression of MAGE-A1 and reactivation of other cancer-testis genes.Cancer Res. 2005 Sep 1;65(17):7751-62. doi: 10.1158/0008-5472.CAN-05-0858. Cancer Res. 2005. PMID: 16140943
-
Hypomethylation of the CTCFL/BORIS promoter and aberrant expression during endometrial cancer progression suggests a role as an Epi-driver gene.Oncotarget. 2014 Feb 28;5(4):1052-61. doi: 10.18632/oncotarget.1697. Oncotarget. 2014. PMID: 24658009 Free PMC article.
-
Discovering a binary CTCF code with a little help from BORIS.Nucleus. 2018 Jan 1;9(1):33-41. doi: 10.1080/19491034.2017.1394536. Epub 2017 Dec 5. Nucleus. 2018. PMID: 29077515 Free PMC article. Review.
-
The novel BORIS + CTCF gene family is uniquely involved in the epigenetics of normal biology and cancer.Semin Cancer Biol. 2002 Oct;12(5):399-414. doi: 10.1016/s1044-579x(02)00060-3. Semin Cancer Biol. 2002. PMID: 12191639 Review.
Cited by
-
Intragenic DNA methylation and BORIS-mediated cancer-specific splicing contribute to the Warburg effect.Proc Natl Acad Sci U S A. 2017 Oct 24;114(43):11440-11445. doi: 10.1073/pnas.1708447114. Epub 2017 Oct 9. Proc Natl Acad Sci U S A. 2017. PMID: 29073069 Free PMC article.
-
Endonuclease-sensitive regions of human spermatozoal chromatin are highly enriched in promoter and CTCF binding sequences.Genome Res. 2009 Aug;19(8):1338-49. doi: 10.1101/gr.094953.109. Epub 2009 Jul 7. Genome Res. 2009. PMID: 19584098 Free PMC article.
-
Exploitation of the interaction of measles virus fusogenic envelope proteins with the surface receptor CD46 on human cells for microcell-mediated chromosome transfer.BMC Biotechnol. 2010 May 6;10:37. doi: 10.1186/1472-6750-10-37. BMC Biotechnol. 2010. PMID: 20444293 Free PMC article.
-
TET-catalyzed oxidation of intragenic 5-methylcytosine regulates CTCF-dependent alternative splicing.EMBO J. 2016 Feb 1;35(3):335-55. doi: 10.15252/embj.201593235. Epub 2015 Dec 28. EMBO J. 2016. PMID: 26711177 Free PMC article.
-
Expression of the epigenetic factor BORIS (CTCFL) in the human genome.J Transl Med. 2011 Dec 14;9:213. doi: 10.1186/1479-5876-9-213. J Transl Med. 2011. PMID: 22168535 Free PMC article. Review.
References
-
- Bell AC, Felsenfeld G. Methylation of a CTCFdependent boundary controls imprinted expression of the Igf2 gene. Nature. 2000;405:482–5. - PubMed
-
- Takai D, Gonzales FA, Tsai YC, Thayer MJ, Jones PA. Large scale mapping of methylcytosines in CTCFbinding sites in the human H19 promoter and aberrant hypomethylation in human bladder cancer. Hum Mol Genet. 2001;10:2619–26. - PubMed
-
- Kanduri C, Pant V, Loukinov D, et al. Functional association of CTCF with the insulator upstreamof the H19 gene is parent of origin-specific and methylationsensitive. Curr Biol. 2000;10:853–6. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

