The RNA-binding proteins HYL1 and SE promote accurate in vitro processing of pri-miRNA by DCL1
- PMID: 18632569
- PMCID: PMC2481344
- DOI: 10.1073/pnas.0803356105
The RNA-binding proteins HYL1 and SE promote accurate in vitro processing of pri-miRNA by DCL1
Abstract
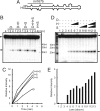
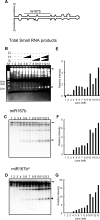
The results of genetic studies in Arabidopsis indicate that three proteins, the RNase III DICER-Like1 (DCL1), the dsRNA-binding protein HYPONASTIC LEAVES1 (HYL1), and the C2H2 Zn-finger protein SERRATE (SE), are required for the accurate processing of microRNA (miRNA) precursors in the plant cell nucleus. To elucidate the biochemical mechanism of miRNA processing, we developed an in vitro miRNA processing assay using purified recombinant proteins. We find that DCL1 alone releases 21-nt short RNAs from dsRNA as well as synthetic miR167b precursor RNAs. However, correctly processed miRNAs constitute a minority of the cleavage products. We show that recombinant HYL1 and SE proteins accelerate the rate of DCL1-mediated cleavage of pre- and pri-miR167b substrates and promote accurate processing.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


Comment in
-
Reconstituting plant miRNA biogenesis.Proc Natl Acad Sci U S A. 2008 Jul 22;105(29):9851-2. doi: 10.1073/pnas.0805207105. Epub 2008 Jul 16. Proc Natl Acad Sci U S A. 2008. PMID: 18632572 Free PMC article. No abstract available.
Similar articles
-
Regulation of miRNA abundance by RNA binding protein TOUGH in Arabidopsis.Proc Natl Acad Sci U S A. 2012 Jul 31;109(31):12817-21. doi: 10.1073/pnas.1204915109. Epub 2012 Jul 16. Proc Natl Acad Sci U S A. 2012. PMID: 22802657 Free PMC article.
-
Dissecting the interactions of SERRATE with RNA and DICER-LIKE 1 in Arabidopsis microRNA precursor processing.Nucleic Acids Res. 2013 Oct;41(19):9129-40. doi: 10.1093/nar/gkt667. Epub 2013 Aug 5. Nucleic Acids Res. 2013. PMID: 23921632 Free PMC article.
-
Homodimerization of HYL1 ensures the correct selection of cleavage sites in primary miRNA.Nucleic Acids Res. 2014 Oct 29;42(19):12224-36. doi: 10.1093/nar/gku907. Epub 2014 Oct 7. Nucleic Acids Res. 2014. PMID: 25294831 Free PMC article.
-
New insights into pri-miRNA processing and accumulation in plants.Wiley Interdiscip Rev RNA. 2015 Sep-Oct;6(5):533-45. doi: 10.1002/wrna.1292. Epub 2015 Jun 29. Wiley Interdiscip Rev RNA. 2015. PMID: 26119101 Review.
-
SERRATE: a key factor in coordinated RNA processing in plants.Trends Plant Sci. 2023 Jul;28(7):841-853. doi: 10.1016/j.tplants.2023.03.009. Epub 2023 Apr 4. Trends Plant Sci. 2023. PMID: 37019716 Review.
Cited by
-
Plant-derived xenomiRs and cancer: Cross-kingdom gene regulation.Saudi J Biol Sci. 2021 Apr;28(4):2408-2422. doi: 10.1016/j.sjbs.2021.01.039. Epub 2021 Feb 1. Saudi J Biol Sci. 2021. PMID: 33911956 Free PMC article. Review.
-
The THO/TREX Complex Active in miRNA Biogenesis Negatively Regulates Root-Associated Acid Phosphatase Activity Induced by Phosphate Starvation.Plant Physiol. 2016 Aug;171(4):2841-53. doi: 10.1104/pp.16.00680. Epub 2016 Jun 21. Plant Physiol. 2016. PMID: 27329222 Free PMC article.
-
microRNA biogenesis and turnover in plants.Cold Spring Harb Symp Quant Biol. 2012;77:183-94. doi: 10.1101/sqb.2013.77.014530. Epub 2013 Feb 25. Cold Spring Harb Symp Quant Biol. 2012. PMID: 23439913 Free PMC article. Review.
-
Capture of regulatory factors via CRISPR-dCas9 for mechanistic analysis of fine-tuned SERRATE expression in Arabidopsis.Nat Plants. 2024 Jan;10(1):86-99. doi: 10.1038/s41477-023-01575-x. Epub 2024 Jan 2. Nat Plants. 2024. PMID: 38168608
-
R-loops at microRNA encoding loci promote co-transcriptional processing of pri-miRNAs in plants.Nat Plants. 2022 Apr;8(4):402-418. doi: 10.1038/s41477-022-01125-x. Epub 2022 Apr 21. Nat Plants. 2022. PMID: 35449404 Free PMC article.
References
-
- Valencia-Sanchez MA, Liu J, Hannon GJ, Parker R. Control of translation and mRNA degradation by miRNAs and siRNAs. Genes Dev. 2006;20:515–524. - PubMed
-
- Pillai RS, Bhattacharyya SN, Filipowicz W. Repression of protein synthesis by miRNAs: How many mechanisms? Trends Cell Biol. 2007;17:118–126. - PubMed
-
- Zhao Y, Srivastava D. A developmental view of microRNA function. Trends Biochem Sci. 2007;32:189–197. - PubMed
-
- Jones-Rhoades MW, Bartel DP, Bartel B. MicroRNAs and their regulatory roles in plants. Annu Rev Plant Biol. 2006;57:19–53. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

