Xist RNA is confined to the nuclear territory of the silenced X chromosome throughout the cell cycle
- PMID: 18625719
- PMCID: PMC2546918
- DOI: 10.1128/MCB.02269-07
Xist RNA is confined to the nuclear territory of the silenced X chromosome throughout the cell cycle
Abstract
In mammalian female cells, one X chromosome is inactivated to prevent a dose difference in the expression of X-encoded proteins between males and females. Xist RNA, required for X chromosome inactivation, is transcribed from the future inactivated X chromosome (Xi), where it spreads in cis, to initiate silencing. We have analyzed Xist RNA transcription and localization throughout the cell cycle. It was found that Xist transcription is constant and that the mature RNA remains attached to the Xi throughout mitosis. Diploid and tetraploid cell lines with an MS2-tagged Xist gene were used to investigate spreading of Xist. Most XXXX(MS2) tetraploid mouse embryonic stem (ES) cells inactivate the X(MS2) chromosome and one other X chromosome. Analysis of cells with two Xi's indicates that Xist RNA is retained by the Xi of its origin and does not spread in trans. Also, in XX(MS2) diploid mouse ES cells with an autosomal Xist transgene, there is no trans exchange of Xist RNA from the Xi to the autosome. We propose that Xist RNA does not dissociate from the Xi of its origin, which precludes a model of diffusion-mediated trans spreading of Xist RNA.
Figures
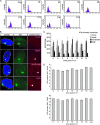
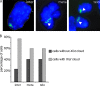
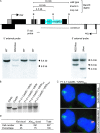
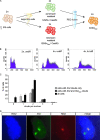
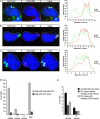
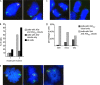
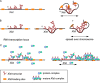
Similar articles
-
Depressing time: Waiting, melancholia, and the psychoanalytic practice of care.In: Kirtsoglou E, Simpson B, editors. The Time of Anthropology: Studies of Contemporary Chronopolitics. Abingdon: Routledge; 2020. Chapter 5. In: Kirtsoglou E, Simpson B, editors. The Time of Anthropology: Studies of Contemporary Chronopolitics. Abingdon: Routledge; 2020. Chapter 5. PMID: 36137063 Free Books & Documents. Review.
-
KDM6A facilitates Xist upregulation at the onset of X inactivation.Biol Sex Differ. 2025 Jan 3;16(1):1. doi: 10.1186/s13293-024-00683-3. Biol Sex Differ. 2025. PMID: 39754175 Free PMC article.
-
Comparison of Two Modern Survival Prediction Tools, SORG-MLA and METSSS, in Patients With Symptomatic Long-bone Metastases Who Underwent Local Treatment With Surgery Followed by Radiotherapy and With Radiotherapy Alone.Clin Orthop Relat Res. 2024 Dec 1;482(12):2193-2208. doi: 10.1097/CORR.0000000000003185. Epub 2024 Jul 23. Clin Orthop Relat Res. 2024. PMID: 39051924
-
Antioxidants for female subfertility.Cochrane Database Syst Rev. 2020 Aug 27;8(8):CD007807. doi: 10.1002/14651858.CD007807.pub4. Cochrane Database Syst Rev. 2020. PMID: 32851663 Free PMC article.
-
Topical fluoride as a cause of dental fluorosis in children.Cochrane Database Syst Rev. 2024 Jun 20;6(6):CD007693. doi: 10.1002/14651858.CD007693.pub3. Cochrane Database Syst Rev. 2024. PMID: 38899538 Review.
Cited by
-
Detection of RNA-DNA association by a proximity ligation-based method.Sci Rep. 2016 Jun 3;6:27313. doi: 10.1038/srep27313. Sci Rep. 2016. PMID: 27256324 Free PMC article.
-
Three-dimensional super-resolution microscopy of the inactive X chromosome territory reveals a collapse of its active nuclear compartment harboring distinct Xist RNA foci.Epigenetics Chromatin. 2014 Apr 28;7:8. doi: 10.1186/1756-8935-7-8. eCollection 2014. Epigenetics Chromatin. 2014. PMID: 25057298 Free PMC article.
-
Subcellular RNA profiling links splicing and nuclear DICER1 to alternative cleavage and polyadenylation.Genome Res. 2016 Jan;26(1):24-35. doi: 10.1101/gr.193995.115. Epub 2015 Nov 6. Genome Res. 2016. PMID: 26546131 Free PMC article.
-
A protein assembly mediates Xist localization and gene silencing.Nature. 2020 Nov;587(7832):145-151. doi: 10.1038/s41586-020-2703-0. Epub 2020 Sep 9. Nature. 2020. PMID: 32908311 Free PMC article.
-
Cohesin is required for higher-order chromatin conformation at the imprinted IGF2-H19 locus.PLoS Genet. 2009 Nov;5(11):e1000739. doi: 10.1371/journal.pgen.1000739. Epub 2009 Nov 26. PLoS Genet. 2009. PMID: 19956766 Free PMC article.
References
-
- Cattanach, B. M., and C. Rasberry. 1994. Identification of the Mus castaneus Xce allele. Mouse Genome 922.
-
- Duthie, S. M., T. B. Nesterova, E. J. Formstone, A. M. Keohane, B. M. Turner, S. M. Zakian, and N. Brockdorff. 1999. Xist RNA exhibits a banded localization on the inactive X chromosome and is excluded from autosomal material in cis. Hum. Mol. Genet. 8195-204. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
