Nuclear import of CaMV P6 is required for infection and suppression of the RNA silencing factor DRB4
- PMID: 18615098
- PMCID: PMC2516879
- DOI: 10.1038/emboj.2008.129
Nuclear import of CaMV P6 is required for infection and suppression of the RNA silencing factor DRB4
Erratum in
-
Nuclear import of CaMV P6 is required for infection and suppression of the RNA silencing factor DRB4.EMBO J. 2015 Oct 14;34(20):2591-2. doi: 10.15252/embj.201570060. Epub 2015 Aug 19. EMBO J. 2015. PMID: 26290338 Free PMC article. No abstract available.
Abstract
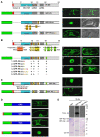
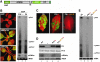
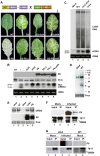
Replication of Cauliflower mosaic virus (CaMV), a plant double-stranded DNA virus, requires the viral translational transactivator protein P6. Although P6 is known to form cytoplasmic inclusion bodies (viroplasms) so far considered essential for virus biology, a fraction of the protein is also present in the nucleus. Here, we report that monomeric P6 is imported into the nucleus through two importin-alpha-dependent nuclear localization signals, and show that this process is mandatory for CaMV infectivity and is independent of translational transactivation and viroplasm formation. One nuclear function of P6 is to suppress RNA silencing, a gene regulation mechanism with antiviral roles, commonly counteracted by dedicated viral suppressor proteins (viral silencing suppressors; VSRs). Transgenic P6 expression in Arabidopsis is genetically equivalent to inactivating the nuclear protein DRB4 that facilitates the activity of the major plant antiviral silencing factor DCL4. We further show that a fraction of P6 immunoprecipitates with DRB4 in CaMV-infected cells. This study identifies both genetic and physical interactions between a VSR to a host RNA silencing component, and highlights the importance of subcellular compartmentalization in VSR function.
Figures


Similar articles
-
Cauliflower mosaic virus protein P6 is a suppressor of RNA silencing.J Gen Virol. 2007 Dec;88(Pt 12):3439-3444. doi: 10.1099/vir.0.83090-0. J Gen Virol. 2007. PMID: 18024914
-
Identification of Strawberry vein banding virus encoded P6 as an RNA silencing suppressor.Virology. 2018 Jul;520:103-110. doi: 10.1016/j.virol.2018.05.003. Epub 2018 May 26. Virology. 2018. PMID: 29843054
-
The open reading frame VI product of Cauliflower mosaic virus is a nucleocytoplasmic protein: its N terminus mediates its nuclear export and formation of electron-dense viroplasms.Plant Cell. 2005 Mar;17(3):927-43. doi: 10.1105/tpc.104.029017. Plant Cell. 2005. PMID: 15746075 Free PMC article.
-
A model for intracellular movement of Cauliflower mosaic virus: the concept of the mobile virion factory.J Exp Bot. 2016 Mar;67(7):2039-48. doi: 10.1093/jxb/erv520. Epub 2015 Dec 18. J Exp Bot. 2016. PMID: 26687180 Review.
-
Setting Up Shop: The Formation and Function of the Viral Factories of Cauliflower mosaic virus.Front Plant Sci. 2017 Oct 30;8:1832. doi: 10.3389/fpls.2017.01832. eCollection 2017. Front Plant Sci. 2017. PMID: 29163571 Free PMC article. Review.
Cited by
-
Cauliflower mosaic virus protein P6 inhibits signaling responses to salicylic acid and regulates innate immunity.PLoS One. 2012;7(10):e47535. doi: 10.1371/journal.pone.0047535. Epub 2012 Oct 11. PLoS One. 2012. PMID: 23071821 Free PMC article.
-
RNA based viral silencing suppression in plant pararetroviruses.Front Plant Sci. 2015 Jun 10;6:398. doi: 10.3389/fpls.2015.00398. eCollection 2015. Front Plant Sci. 2015. PMID: 26113850 Free PMC article. Review.
-
Importin α2 participates in RNA interference against bamboo mosaic virus accumulation in Nicotiana benthamiana via NbAGO10a-mediated small RNA clearance.Mol Plant Pathol. 2024 Jan;25(1):e13422. doi: 10.1111/mpp.13422. Mol Plant Pathol. 2024. PMID: 38279848 Free PMC article.
-
Identification of the Potential Virulence Factors and RNA Silencing Suppressors of Mulberry Mosaic Dwarf-Associated Geminivirus.Viruses. 2018 Sep 3;10(9):472. doi: 10.3390/v10090472. Viruses. 2018. PMID: 30177616 Free PMC article.
-
Identification of Silencing Suppressor Protein Encoded by Strawberry Mottle Virus.Front Plant Sci. 2022 May 31;13:786489. doi: 10.3389/fpls.2022.786489. eCollection 2022. Front Plant Sci. 2022. PMID: 35712581 Free PMC article.
References
-
- Adenot X, Elmayan T, Lauressergues D, Boutet S, Bouche N, Gasciolli V, Vaucheret H (2006) DRB4-dependent TAS3 trans-acting siRNAs control leaf morphology through AGO7. Curr Biol 16: 927–932 - PubMed
-
- Al-Kaff NS, Covey SN, Kreike MM, Page AM, Pinder R, Dale PJ (1998) Transcriptional and posttranscriptional plant gene silencing in response to a pathogen. Science 279: 2113–2115 - PubMed
-
- Baumberger N, Tsai CH, Lie M, Havecker E, Baulcombe DC (2007) The polerovirus silencing suppressor P0 targets ARGONAUTE proteins for degradation. Curr Biol 17: 1609–1614 - PubMed
-
- Bonneville JM, Sanfaçon H, Futterer J, Hohn T (1989) Posttranscriptional trans-activation in cauliflower mosaic virus. Cell 59: 1135–1143 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

