Altered gene expression changes in Arabidopsis leaf tissues and protoplasts in response to Plum pox virus infection
- PMID: 18613973
- PMCID: PMC2478689
- DOI: 10.1186/1471-2164-9-325
Altered gene expression changes in Arabidopsis leaf tissues and protoplasts in response to Plum pox virus infection
Abstract
Background: Virus infection induces the activation and suppression of global gene expression in the host. Profiling gene expression changes in the host may provide insights into the molecular mechanisms that underlie host physiological and phenotypic responses to virus infection. In this study, the Arabidopsis Affymetrix ATH1 array was used to assess global gene expression changes in Arabidopsis thaliana plants infected with Plum pox virus (PPV). To identify early genes in response to PPV infection, an Arabidopsis synchronized single-cell transformation system was developed. Arabidopsis protoplasts were transfected with a PPV infectious clone and global gene expression changes in the transfected protoplasts were profiled.
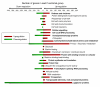
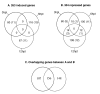
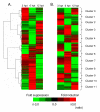
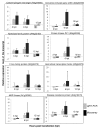
Results: Microarray analysis of PPV-infected Arabidopsis leaf tissues identified 2013 and 1457 genes that were significantly (Q < or = 0.05) up- (> or = 2.5 fold) and downregulated (< or = -2.5 fold), respectively. Genes associated with soluble sugar, starch and amino acid, intracellular membrane/membrane-bound organelles, chloroplast, and protein fate were upregulated, while genes related to development/storage proteins, protein synthesis and translation, and cell wall-associated components were downregulated. These gene expression changes were associated with PPV infection and symptom development. Further transcriptional profiling of protoplasts transfected with a PPV infectious clone revealed the upregulation of defence and cellular signalling genes as early as 6 hours post transfection. A cross sequence comparison analysis of genes differentially regulated by PPV-infected Arabidopsis leaves against uniEST sequences derived from PPV-infected leaves of Prunus persica, a natural host of PPV, identified orthologs related to defence, metabolism and protein synthesis. The cross comparison of genes differentially regulated by PPV infection and by the infections of other positive sense RNA viruses revealed a common set of 416 genes. These identified genes, particularly the early responsive genes, may be critical in virus infection.
Conclusion: Gene expression changes in PPV-infected Arabidopsis are the molecular basis of stress and defence-like responses, PPV pathogenesis and symptom development. The differentially regulated genes, particularly the early responsive genes, and a common set of genes regulated by infections of PPV and other positive sense RNA viruses identified in this study are candidates suitable for further functional characterization to shed lights on molecular virus-host interactions.
Figures


Similar articles
-
Transfection of Arabidopsis protoplasts with a Plum pox virus (PPV) infectious clone for studying early molecular events associated with PPV infection.J Virol Methods. 2006 Sep;136(1-2):147-53. doi: 10.1016/j.jviromet.2006.05.009. Epub 2006 Jun 13. J Virol Methods. 2006. PMID: 16777241
-
Allelic variation at the rpv1 locus controls partial resistance to Plum pox virus infection in Arabidopsis thaliana.BMC Plant Biol. 2015 Jun 25;15:159. doi: 10.1186/s12870-015-0559-5. BMC Plant Biol. 2015. PMID: 26109391 Free PMC article.
-
Gene Expression Analysis of Plum pox virus (Sharka) Susceptibility/Resistance in Apricot (Prunus armeniaca L.).PLoS One. 2015 Dec 11;10(12):e0144670. doi: 10.1371/journal.pone.0144670. eCollection 2015. PLoS One. 2015. PMID: 26658051 Free PMC article.
-
Sharka: how do plants respond to Plum pox virus infection?J Exp Bot. 2015 Jan;66(1):25-35. doi: 10.1093/jxb/eru428. Epub 2014 Oct 21. J Exp Bot. 2015. PMID: 25336685 Review.
-
The use of transgenic fruit trees as a resistance strategy for virus epidemics: the plum pox (sharka) model.Virus Res. 2000 Nov;71(1-2):63-9. doi: 10.1016/s0168-1702(00)00188-x. Virus Res. 2000. PMID: 11137162 Review.
Cited by
-
Resistance to Plum Pox Virus (PPV) in apricot (Prunus armeniaca L.) is associated with down-regulation of two MATHd genes.BMC Plant Biol. 2018 Jan 27;18(1):25. doi: 10.1186/s12870-018-1237-1. BMC Plant Biol. 2018. PMID: 29374454 Free PMC article.
-
Proteolytic Processing of Plant Proteins by Potyvirus NIa Proteases.J Virol. 2022 Jan 26;96(2):e0144421. doi: 10.1128/JVI.01444-21. Epub 2021 Nov 10. J Virol. 2022. PMID: 34757836 Free PMC article.
-
Relationship between symptoms and gene expression induced by the infection of three strains of Rice dwarf virus.PLoS One. 2011 Mar 22;6(3):e18094. doi: 10.1371/journal.pone.0018094. PLoS One. 2011. PMID: 21445363 Free PMC article.
-
Identification and functional characterization of NbMLP28, a novel MLP-like protein 28 enhancing Potato virus Y resistance in Nicotiana benthamiana.BMC Microbiol. 2020 Mar 6;20(1):55. doi: 10.1186/s12866-020-01725-7. BMC Microbiol. 2020. PMID: 32143563 Free PMC article.
-
Transcriptomic analysis of Prunus domestica undergoing hypersensitive response to plum pox virus infection.PLoS One. 2014 Jun 24;9(6):e100477. doi: 10.1371/journal.pone.0100477. eCollection 2014. PLoS One. 2014. PMID: 24959894 Free PMC article.
References
-
- Maule A, Leh V, Lederer C. The dialogue between viruses and hosts in compatible interactions. Curr Opin Plant Biol. 2002;5:279–284. - PubMed
-
- Whitham SA, Yang C, Goodin MM. Global impact: elucidating plant responses to viral infection. Mol Plant Microbe Interact. 2006;19:1207–1215. - PubMed
-
- Stange C. Plant-virus interactions during the infective process. Cien Inv Agr. 2006;33:1–18.
-
- Culver JN, Padmanabhan MS. Virus-induced disease: Altering host physiology one interaction at a time. Annu Rev Phytopathol. 2007;45:221–243. - PubMed
-
- Whitham SA, Wang Y. Roles for host factors in plant viral pathogenicity. Curr Opin Plant Biol. 2004;7:365–371. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

