Molecular ecology and natural history of simian foamy virus infection in wild-living chimpanzees
- PMID: 18604273
- PMCID: PMC2435277
- DOI: 10.1371/journal.ppat.1000097
Molecular ecology and natural history of simian foamy virus infection in wild-living chimpanzees
Abstract
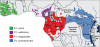
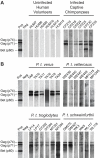
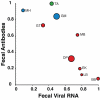
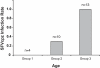
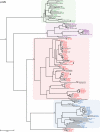
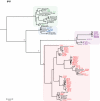
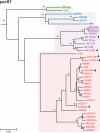
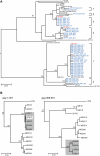
Identifying microbial pathogens with zoonotic potential in wild-living primates can be important to human health, as evidenced by human immunodeficiency viruses types 1 and 2 (HIV-1 and HIV-2) and Ebola virus. Simian foamy viruses (SFVs) are ancient retroviruses that infect Old and New World monkeys and apes. Although not known to cause disease, these viruses are of public health interest because they have the potential to infect humans and thus provide a more general indication of zoonotic exposure risks. Surprisingly, no information exists concerning the prevalence, geographic distribution, and genetic diversity of SFVs in wild-living monkeys and apes. Here, we report the first comprehensive survey of SFVcpz infection in free-ranging chimpanzees (Pan troglodytes) using newly developed, fecal-based assays. Chimpanzee fecal samples (n = 724) were collected at 25 field sites throughout equatorial Africa and tested for SFVcpz-specific antibodies (n = 706) or viral nucleic acids (n = 392). SFVcpz infection was documented at all field sites, with prevalence rates ranging from 44% to 100%. In two habituated communities, adult chimpanzees had significantly higher SFVcpz infection rates than infants and juveniles, indicating predominantly horizontal rather than vertical transmission routes. Some chimpanzees were co-infected with simian immunodeficiency virus (SIVcpz); however, there was no evidence that SFVcpz and SIVcpz were epidemiologically linked. SFVcpz nucleic acids were recovered from 177 fecal samples, all of which contained SFVcpz RNA and not DNA. Phylogenetic analysis of partial gag (616 bp), pol-RT (717 bp), and pol-IN (425 bp) sequences identified a diverse group of viruses, which could be subdivided into four distinct SFVcpz lineages according to their chimpanzee subspecies of origin. Within these lineages, there was evidence of frequent superinfection and viral recombination. One chimpanzee was infected by a foamy virus from a Cercopithecus monkey species, indicating cross-species transmission of SFVs in the wild. These data indicate that SFVcpz (i) is widely distributed among all chimpanzee subspecies; (ii) is shed in fecal samples as viral RNA; (iii) is transmitted predominantly by horizontal routes; (iv) is prone to superinfection and recombination; (v) has co-evolved with its natural host; and (vi) represents a sensitive marker of population structure that may be useful for chimpanzee taxonomy and conservation strategies.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures

Similar articles
-
Detection and molecular characterization of foamy viruses in Central African chimpanzees of the Pan troglodytes troglodytes and Pan troglodytes vellerosus subspecies.J Med Primatol. 2006 Apr;35(2):59-66. doi: 10.1111/j.1600-0684.2006.00149.x. J Med Primatol. 2006. PMID: 16556292
-
Origin of HIV-1 in the chimpanzee Pan troglodytes troglodytes.Nature. 1999 Feb 4;397(6718):436-41. doi: 10.1038/17130. Nature. 1999. PMID: 9989410
-
Cocirculation of Two env Molecular Variants, of Possible Recombinant Origin, in Gorilla and Chimpanzee Simian Foamy Virus Strains from Central Africa.J Virol. 2015 Dec;89(24):12480-91. doi: 10.1128/JVI.01798-15. Epub 2015 Oct 7. J Virol. 2015. PMID: 26446599 Free PMC article.
-
[Mechanisms of viral emergence and interspecies transmission: the exemple of simian foamy viruses in Central Africa].Bull Acad Natl Med. 2013 Dec;197(9):1655-67; discussion 1667-8. doi: 10.1016/S0001-4079(19)31387-1. Bull Acad Natl Med. 2013. PMID: 26137812 Free PMC article. Review. French.
-
Simian retroviruses in African apes.Clin Microbiol Infect. 2012 Jun;18(6):514-20. doi: 10.1111/j.1469-0691.2012.03843.x. Epub 2012 Apr 20. Clin Microbiol Infect. 2012. PMID: 22515409 Review.
Cited by
-
Multiple Infiltration and Cross-Species Transmission of Foamy Viruses across the Paleozoic to the Cenozoic Era.J Virol. 2021 Jun 24;95(14):e0048421. doi: 10.1128/JVI.00484-21. Epub 2021 Jun 24. J Virol. 2021. PMID: 33910951 Free PMC article.
-
HTLV-3/4 and simian foamy retroviruses in humans: discovery, epidemiology, cross-species transmission and molecular virology.Virology. 2013 Jan 5;435(1):187-99. doi: 10.1016/j.virol.2012.09.035. Virology. 2013. PMID: 23217627 Free PMC article. Review.
-
Identification of recombination in the envelope gene of simian foamy virus serotype 2 isolated from Macaca cyclopis.J Virol. 2013 Aug;87(15):8792-7. doi: 10.1128/JVI.03555-12. Epub 2013 May 22. J Virol. 2013. PMID: 23698303 Free PMC article.
-
Molecular biology of foamy viruses.Med Microbiol Immunol. 2010 Aug;199(3):197-207. doi: 10.1007/s00430-010-0158-x. Epub 2010 May 6. Med Microbiol Immunol. 2010. PMID: 20445989 Review.
-
High prevalence of simian immunodeficiency virus infection in a community of savanna chimpanzees.J Virol. 2011 Oct;85(19):9918-28. doi: 10.1128/JVI.05475-11. Epub 2011 Jul 20. J Virol. 2011. PMID: 21775446 Free PMC article.
References
-
- Delelis O, Lehmann-Che J, Saib A. Foamy viruses–a world apart. Curr Opin Microbiol. 2004;7:400–406. - PubMed
-
- Murray SM, Linial ML. Foamy virus infection in primates. J Med Primatol. 2006;35:225–235. - PubMed
-
- Rethwilm A. Foamy Viruses. In: Mahy BWJ, ter Meulen V, editors. Topley & Wilson's Microbiology and Microbial Infections-Virology. London: Hodder Arnold; 2005. pp. 1304–1321.
-
- Broussard SR, Comuzzie AG, Leighton KL, Leland MM, Whitehead EM, et al. Characterization of new simian foamy viruses from African nonhuman primates. Virology. 1997;237:349–359. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

