APOBEC3G and APOBEC3F require an endogenous cofactor to block HIV-1 replication
- PMID: 18604271
- PMCID: PMC2435275
- DOI: 10.1371/journal.ppat.1000095
APOBEC3G and APOBEC3F require an endogenous cofactor to block HIV-1 replication
Abstract
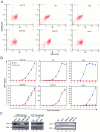
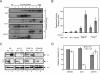
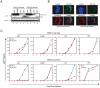
APOBEC3G (A3G)/APOBEC3F (A3F) are two members of APOBEC3 cytidine deaminase subfamily. Although they potently inhibit the replication of vif-deficient HIV-1, this mechanism is still poorly understood. Initially, A3G/A3F were thought to catalyze C-to-U transitions on the minus-strand viral cDNAs during reverse transcription to disrupt the viral life cycle. Recently, it was found more likely that A3G/A3F directly interrupts viral reverse transcription or integration. In addition, A3G/A3F are both found in the high-molecular-mass complex in immortalized cell lines, where they interact with a number of different cellular proteins. However, there has been no evidence to prove that these interactions are required for A3G/A3F function. Here, we studied A3G/A3F-restricted HIV-1 replication in six different human T cell lines by infecting them with wild-type or vif-deficient HIV-1. Interestingly, in a CEM-derived cell line CEM-T4, which expresses high levels of A3G/A3F proteins, the vif-deficient virus replicated as equally well as the wild-type virus, suggesting that these endogenous antiretroviral genes lost anti-HIV activities. It was confirmed that these A3G/A3F genes do not contain any mutation and are functionally normal. Consistently, overexpression of exogenous A3G/A3F in CEM-T4 cells still failed to restore their anti-HIV activities. However, this activity could be restored if CEM-T4 cells were fused to 293T cells to form heterokaryons. These results demonstrate that CEM-T4 cells lack a cellular cofactor, which is critical for A3G/A3F anti-HIV activity. We propose that a further study of this novel factor will provide another strategy for a complete understanding of the A3G/A3F antiretroviral mechanism.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures


Similar articles
-
APOBEC3G and APOBEC3F Act in Concert To Extinguish HIV-1 Replication.J Virol. 2016 Apr 14;90(9):4681-4695. doi: 10.1128/JVI.03275-15. Print 2016 May. J Virol. 2016. PMID: 26912618 Free PMC article.
-
Distinct domains within APOBEC3G and APOBEC3F interact with separate regions of human immunodeficiency virus type 1 Vif.J Virol. 2009 Feb;83(4):1992-2003. doi: 10.1128/JVI.01621-08. Epub 2008 Nov 26. J Virol. 2009. PMID: 19036809 Free PMC article.
-
Identification of two distinct human immunodeficiency virus type 1 Vif determinants critical for interactions with human APOBEC3G and APOBEC3F.J Virol. 2007 Aug;81(15):8201-10. doi: 10.1128/JVI.00395-07. Epub 2007 May 23. J Virol. 2007. PMID: 17522216 Free PMC article.
-
Various strategies for developing APOBEC3G protectors to circumvent human immunodeficiency virus type 1.Eur J Med Chem. 2023 Mar 15;250:115188. doi: 10.1016/j.ejmech.2023.115188. Epub 2023 Feb 6. Eur J Med Chem. 2023. PMID: 36773550 Review.
-
Multiple ways of targeting APOBEC3-virion infectivity factor interactions for anti-HIV-1 drug development.Trends Pharmacol Sci. 2009 Dec;30(12):638-46. doi: 10.1016/j.tips.2009.09.006. Trends Pharmacol Sci. 2009. PMID: 19837465 Free PMC article. Review.
Cited by
-
Multiple Inhibitory Factors Act in the Late Phase of HIV-1 Replication: a Systematic Review of the Literature.Microbiol Mol Biol Rev. 2018 Jan 10;82(1):e00051-17. doi: 10.1128/MMBR.00051-17. Print 2018 Mar. Microbiol Mol Biol Rev. 2018. PMID: 29321222 Free PMC article. Review.
-
CEM-T4 cells do not lack an APOBEC3G cofactor.PLoS Pathog. 2009 Jul;5(7):e1000528. doi: 10.1371/journal.ppat.1000528. Epub 2009 Jul 31. PLoS Pathog. 2009. PMID: 19649317 Free PMC article. No abstract available.
-
Interactions of host APOBEC3 restriction factors with HIV-1 in vivo: implications for therapeutics.Expert Rev Mol Med. 2010 Jan 22;12:e4. doi: 10.1017/S1462399409001343. Expert Rev Mol Med. 2010. PMID: 20096141 Free PMC article. Review.
-
Catalytic activity of APOBEC3F is required for efficient restriction of Vif-deficient human immunodeficiency virus.Virology. 2014 Feb;450-451:49-54. doi: 10.1016/j.virol.2013.11.041. Epub 2013 Dec 20. Virology. 2014. PMID: 24503066 Free PMC article.
-
Crystal structure of the DNA cytosine deaminase APOBEC3F: the catalytically active and HIV-1 Vif-binding domain.Structure. 2013 Jun 4;21(6):1042-50. doi: 10.1016/j.str.2013.04.010. Epub 2013 May 16. Structure. 2013. PMID: 23685212 Free PMC article.
References
-
- Holmes RK, Malim MH, Bishop KN. APOBEC-mediated viral restriction: Not simply editing? Trends Biochem Sci. 2007;32:118–128. - PubMed
-
- Teng B, Burant CF, Davidson NO. Molecular cloning of an apolipoprotein B messenger RNA editing protein. Science. 1993;260:1816–1819. - PubMed
-
- Muramatsu M, Sankaranand VS, Anant S, Sugai M, Kinoshita K, et al. Specific expression of activation-induced cytidine deaminase (AID), a novel member of the RNA-editing deaminase family in germinal center B cells. J Biol Chem. 1999;274:18470–18476. - PubMed
-
- Bishop KN, Holmes RK, Sheehy AM, Davidson NO, Cho SJ, et al. Cytidine deamination of retroviral DNA by diverse APOBEC proteins. Curr Biol. 2004;14:1392–1396. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

