Mre11-Rad50-Nbs1-dependent processing of DNA breaks generates oligonucleotides that stimulate ATM activity
- PMID: 18596698
- PMCID: PMC2453060
- DOI: 10.1038/emboj.2008.128
Mre11-Rad50-Nbs1-dependent processing of DNA breaks generates oligonucleotides that stimulate ATM activity
Abstract
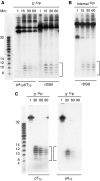
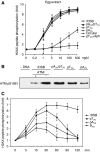
DNA double-strand breaks (DSBs) can be processed by the Mre11-Rad50-Nbs1 (MRN) complex, which is essential to promote ataxia telangiectasia-mutated (ATM) activation. However, the molecular mechanisms linking MRN activity to ATM are not fully understood. Here, using Xenopus laevis egg extract we show that MRN-dependent processing of DSBs leads to the accumulation of short single-stranded DNA oligonucleotides (ssDNA oligos). The MRN complex isolated from the extract containing DSBs is bound to ssDNA oligos and stimulates ATM activity. Elimination of ssDNA oligos results in rapid extinction of ATM activity. Significantly, ssDNA oligos can be isolated from human cells damaged with ionizing radiation and injection of small synthetic ssDNA oligos into undamaged cells also induces ATM activation. These results suggest that MRN-dependent generation of ssDNA oligos, which constitute a unique signal of ongoing DSB repair not encountered in normal DNA metabolism, stimulates ATM activity.
Figures
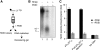
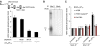

Similar articles
-
Ataxia telangiectasia-mutated (ATM) kinase activity is regulated by ATP-driven conformational changes in the Mre11/Rad50/Nbs1 (MRN) complex.J Biol Chem. 2013 May 3;288(18):12840-51. doi: 10.1074/jbc.M113.460378. Epub 2013 Mar 22. J Biol Chem. 2013. PMID: 23525106 Free PMC article.
-
The Mre11/Rad50/Nbs1 complex and its role as a DNA double-strand break sensor for ATM.Cell Cycle. 2005 Jun;4(6):737-40. doi: 10.4161/cc.4.6.1715. Epub 2005 Jun 6. Cell Cycle. 2005. PMID: 15908798
-
Mre11-Rad50-Nbs1 is a keystone complex connecting DNA repair machinery, double-strand break signaling, and the chromatin template.Biochem Cell Biol. 2007 Aug;85(4):509-20. doi: 10.1139/O07-069. Biochem Cell Biol. 2007. PMID: 17713585 Review.
-
ATM activation by DNA double-strand breaks through the Mre11-Rad50-Nbs1 complex.Science. 2005 Apr 22;308(5721):551-4. doi: 10.1126/science.1108297. Epub 2005 Mar 24. Science. 2005. PMID: 15790808
-
Activation and regulation of ATM kinase activity in response to DNA double-strand breaks.Oncogene. 2007 Dec 10;26(56):7741-8. doi: 10.1038/sj.onc.1210872. Oncogene. 2007. PMID: 18066086 Review.
Cited by
-
Functional interplay between the oxidative stress response and DNA damage checkpoint signaling for genome maintenance in aerobic organisms.J Microbiol. 2020 Feb;58(2):81-91. doi: 10.1007/s12275-020-9520-x. Epub 2019 Dec 23. J Microbiol. 2020. PMID: 31875928 Review.
-
DNA resection in eukaryotes: deciding how to fix the break.Nat Struct Mol Biol. 2010 Jan;17(1):11-6. doi: 10.1038/nsmb.1710. Nat Struct Mol Biol. 2010. PMID: 20051983 Free PMC article. Review.
-
Intracellular traffic of oligodeoxynucleotides in and out of the nucleus: effect of exportins and DNA structure.Oligonucleotides. 2010 Dec;20(6):277-84. doi: 10.1089/oli.2010.0255. Epub 2010 Oct 14. Oligonucleotides. 2010. PMID: 20946012 Free PMC article.
-
Activation of the DNA-repair mechanism through NBS1 and MRE11 diffusion.PLoS Comput Biol. 2018 Jul 27;14(7):e1006362. doi: 10.1371/journal.pcbi.1006362. eCollection 2018 Jul. PLoS Comput Biol. 2018. PMID: 30052627 Free PMC article.
-
DNA damage response in renal ischemia-reperfusion and ATP-depletion injury of renal tubular cells.Biochim Biophys Acta. 2014 Jul;1842(7):1088-96. doi: 10.1016/j.bbadis.2014.04.002. Epub 2014 Apr 12. Biochim Biophys Acta. 2014. PMID: 24726884 Free PMC article.
References
-
- Bakkenist CJ, Kastan MB (2003) DNA damage activates ATM through intermolecular autophosphorylation and dimer dissociation. Nature 421: 499–506 - PubMed
-
- Berkovich E, Monnat RJ Jr, Kastan MB (2007) Roles of ATM and NBS1 in chromatin structure modulation and DNA double-strand break repair. Nat Cell Biol 9: 683–690 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

