Regulation of protein translation through mRNA structure influences MHC class I loading and T cell recognition
- PMID: 18591662
- PMCID: PMC2453702
- DOI: 10.1073/pnas.0801968105
Regulation of protein translation through mRNA structure influences MHC class I loading and T cell recognition
Abstract
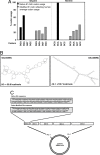
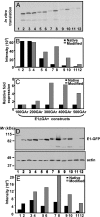
Many viruses avoid immune surveillance during latent infection through reduction in the synthesis of virally encoded proteins. Although antigen presentation critically depends on the level of viral protein synthesis, the precise mechanism used to regulate the generation of antigenic peptide precursors remains elusive. Here, we demonstrate that a purine overloaded virally encoded mRNA lacking secondary structure significantly impacts the efficiency of protein translation and prevents endogenous antigen presentation. Reducing this purine bias through the generation of constructs expressing codon-modified sequences, while maintaining the encoded protein sequence, increased the stem-loop structure of the corresponding mRNA and dramatically enhanced self-synthesis of the viral protein. As a consequence, a higher number of HLA-peptide complexes were detected on the surface of cells expressing this viral protein. Furthermore, these cells were more efficiently recognized by virus-specific T cells compared with those expressing the same antigen expressed by a purine-biased mRNA. These findings delineate a mechanism by which viruses regulate self-synthesis of proteins and offer an effective strategy to evade CD8(+) T cell-mediated immune regulation.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

Comment in
-
Immune surveillance obstructed by viral mRNA.Proc Natl Acad Sci U S A. 2008 Jul 8;105(27):9135-6. doi: 10.1073/pnas.0804456105. Epub 2008 Jul 1. Proc Natl Acad Sci U S A. 2008. PMID: 18599434 Free PMC article. No abstract available.
Similar articles
-
mRNA Structural constraints on EBNA1 synthesis impact on in vivo antigen presentation and early priming of CD8+ T cells.PLoS Pathog. 2014 Oct 9;10(10):e1004423. doi: 10.1371/journal.ppat.1004423. eCollection 2014 Oct. PLoS Pathog. 2014. PMID: 25299404 Free PMC article.
-
Epstein Barr virus-encoded EBNA1 interference with MHC class I antigen presentation reveals a close correlation between mRNA translation initiation and antigen presentation.PLoS Pathog. 2010 Oct 14;6(10):e1001151. doi: 10.1371/journal.ppat.1001151. PLoS Pathog. 2010. PMID: 20976201 Free PMC article.
-
Messenger RNA sequence rather than protein sequence determines the level of self-synthesis and antigen presentation of the EBV-encoded antigen, EBNA1.PLoS Pathog. 2012 Dec;8(12):e1003112. doi: 10.1371/journal.ppat.1003112. Epub 2012 Dec 27. PLoS Pathog. 2012. PMID: 23300450 Free PMC article.
-
Effects of messenger RNA structure and other translational control mechanisms on major histocompatibility complex-I mediated antigen presentation.Wiley Interdiscip Rev RNA. 2015 Mar-Apr;6(2):157-71. doi: 10.1002/wrna.1262. Epub 2014 Sep 26. Wiley Interdiscip Rev RNA. 2015. PMID: 25264139 Free PMC article. Review.
-
Epstein-Barr virus evasion of CD8(+) and CD4(+) T cell immunity via concerted actions of multiple gene products.Semin Cancer Biol. 2008 Dec;18(6):397-408. doi: 10.1016/j.semcancer.2008.10.008. Epub 2008 Oct 25. Semin Cancer Biol. 2008. PMID: 18977445 Review.
Cited by
-
Host translation at the nexus of infection and immunity.Cell Host Microbe. 2012 Oct 18;12(4):470-83. doi: 10.1016/j.chom.2012.09.006. Cell Host Microbe. 2012. PMID: 23084916 Free PMC article. Review.
-
Autophagy and immunity - insights from human herpesviruses.Front Immunol. 2012 Jul 4;3:170. doi: 10.3389/fimmu.2012.00170. eCollection 2012. Front Immunol. 2012. PMID: 22783253 Free PMC article.
-
T cell detection of a B-cell tropic virus infection: newly-synthesised versus mature viral proteins as antigen sources for CD4 and CD8 epitope display.PLoS Pathog. 2009 Dec;5(12):e1000699. doi: 10.1371/journal.ppat.1000699. Epub 2009 Dec 18. PLoS Pathog. 2009. PMID: 20019813 Free PMC article.
-
mRNA translation regulation by the Gly-Ala repeat of Epstein-Barr virus nuclear antigen 1.J Virol. 2009 Feb;83(3):1289-98. doi: 10.1128/JVI.01369-08. Epub 2008 Nov 19. J Virol. 2009. PMID: 19019958 Free PMC article.
-
Monitoring peptide processing for MHC class I molecules in the endoplasmic reticulum.Curr Opin Immunol. 2014 Feb;26:123-7. doi: 10.1016/j.coi.2013.11.006. Epub 2013 Dec 11. Curr Opin Immunol. 2014. PMID: 24556408 Free PMC article. Review.
References
-
- Mocarski ES., Jr Immunomodulation by cytomegaloviruses: Manipulative strategies beyond evasion. Trends Microbiol. 2002;10:332–339. - PubMed
-
- Khanna R, et al. EBV peptide epitope sensitization restores human cytotoxic T cell recognition of Burkitt's lymphoma cells. Evidence for a critical role for ICAM-2. J Immunol. 1993;150:5154–5162. - PubMed
-
- Gandhi MK, Khanna R. Human cytomegalovirus: Clinical aspects, immune regulation, and emerging treatments. Lancet Infect Dis. 2004;4:725–738. - PubMed
-
- Khanna R, Moss DJ, Gandhi M. Applications of emerging immunotherapeutic strategies for Epstein–Barr virus-associated malignancies. Nat Clin Pract Oncol. 2005;2:138–149. - PubMed
-
- Bauer D, Tampe R. Herpes viral proteins blocking the transporter associated with antigen processing TAP: From genes to function and structure. Curr Top Microbiol Immunol. 2002;269:87–99. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

