An activity-regulated microRNA controls dendritic plasticity by down-regulating p250GAP
- PMID: 18577589
- PMCID: PMC2449370
- DOI: 10.1073/pnas.0803072105
An activity-regulated microRNA controls dendritic plasticity by down-regulating p250GAP
Abstract
Activity-regulated gene expression is believed to play a key role in the development and refinement of neuronal circuitry. Nevertheless, the transcriptional networks that regulate synapse growth and plasticity remain largely uncharacterized. Here, we show that microRNA 132 (miR132) is an activity-dependent rapid response gene regulated by the cAMP response element-binding (CREB) protein pathway. Introduction of miR132 into hippocampal neurons enhanced dendrite morphogenesis whereas inhibition of miR132 by 2'O-methyl RNA antagonists blocked these effects. Furthermore, neuronal activity inhibited translation of p250GAP, a miR132 target, and siRNA-mediated knockdown of p250GAP mimicked miR132-induced dendrite growth. Experiments using dominant-interfering mutants suggested that Rac signaling is downstream of miR132 and p250GAP. We propose that the miR132-p250GAP pathway plays a key role in activity-dependent structural and functional plasticity.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
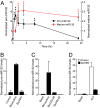
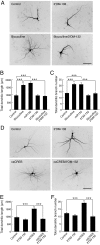
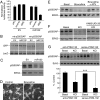
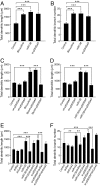
Similar articles
-
An activity-induced microRNA controls dendritic spine formation by regulating Rac1-PAK signaling.Mol Cell Neurosci. 2010 Jan;43(1):146-56. doi: 10.1016/j.mcn.2009.10.005. Epub 2009 Oct 20. Mol Cell Neurosci. 2010. PMID: 19850129 Free PMC article.
-
Leptin induces hippocampal synaptogenesis via CREB-regulated microRNA-132 suppression of p250GAP.Mol Endocrinol. 2014 Jul;28(7):1073-87. doi: 10.1210/me.2013-1332. Epub 2014 May 30. Mol Endocrinol. 2014. PMID: 24877561 Free PMC article.
-
A cAMP-response element binding protein-induced microRNA regulates neuronal morphogenesis.Proc Natl Acad Sci U S A. 2005 Nov 8;102(45):16426-31. doi: 10.1073/pnas.0508448102. Epub 2005 Oct 31. Proc Natl Acad Sci U S A. 2005. PMID: 16260724 Free PMC article.
-
Cataloguing and Selection of mRNAs Localized to Dendrites in Neurons and Regulated by RNA-Binding Proteins in RNA Granules.Biomolecules. 2020 Jan 22;10(2):167. doi: 10.3390/biom10020167. Biomolecules. 2020. PMID: 31978946 Free PMC article. Review.
-
Synaptic plasticity: regulated translation in dendrites.Curr Biol. 1999 Mar 11;9(5):R168-70. doi: 10.1016/s0960-9822(99)80104-3. Curr Biol. 1999. PMID: 10074442 Review.
Cited by
-
MicroRNA-132 Interact with p250GAP/Cdc42 Pathway in the Hippocampal Neuronal Culture Model of Acquired Epilepsy and Associated with Epileptogenesis Process.Neural Plast. 2016;2016:5108489. doi: 10.1155/2016/5108489. Epub 2016 Aug 8. Neural Plast. 2016. PMID: 27579184 Free PMC article.
-
Luteolin induces microRNA-132 expression and modulates neurite outgrowth in PC12 cells.PLoS One. 2012;7(8):e43304. doi: 10.1371/journal.pone.0043304. Epub 2012 Aug 16. PLoS One. 2012. PMID: 22916239 Free PMC article.
-
Transcript specificity in BDNF-regulated protein synthesis.Neuropharmacology. 2014 Jan;76 Pt C(0 0):657-63. doi: 10.1016/j.neuropharm.2013.05.004. Epub 2013 May 22. Neuropharmacology. 2014. PMID: 23707639 Free PMC article. Review.
-
miR-9 and miR-124 synergistically affect regulation of dendritic branching via the AKT/GSK3β pathway by targeting Rap2a.Sci Rep. 2016 May 25;6:26781. doi: 10.1038/srep26781. Sci Rep. 2016. PMID: 27221778 Free PMC article.
-
miR-132/212 is induced by stress and its dysregulation triggers anxiety-related behavior.Neuropharmacology. 2019 Jan;144:256-270. doi: 10.1016/j.neuropharm.2018.10.020. Epub 2018 Oct 18. Neuropharmacology. 2019. PMID: 30342060 Free PMC article.
References
-
- Katz LC, Shatz CJ. Synaptic activity and the construction of cortical circuits. Science. 1996;274:1133–1138. - PubMed
-
- Yuste R, Bonhoeffer T. Morphological changes in dendritic spines associated with long-term synaptic plasticity. Annu Rev Neurosci. 2001;24:1071–1089. - PubMed
-
- Wong RO, Ghosh A. Activity-dependent regulation of dendritic growth and patterning. Nat Rev Neurosci. 2002;3:803–812. - PubMed
-
- Redmond L, Kashani AH, Ghosh A. Calcium regulation of dendritic growth via CaM kinase IV and CREB-mediated transcription. Neuron. 2002;34:999–1010. - PubMed
-
- Wayman GA, et al. Activity-dependent dendritic arborization mediated by CaM-kinase I activation and enhanced CREB-dependent transcription of Wnt-2. Neuron. 2006;50:897–909. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

