Sumo-1 function is dispensable in normal mouse development
- PMID: 18573887
- PMCID: PMC2519746
- DOI: 10.1128/MCB.00651-08
Sumo-1 function is dispensable in normal mouse development
Abstract
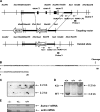
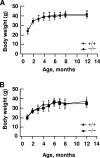
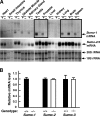
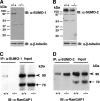
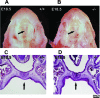
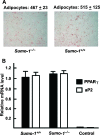
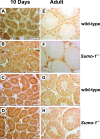
To elucidate SUMO-1 functions in vivo, we targeted by homologous recombination the last three exons of the murine Sumo-1 gene. Sumo-1 mRNA abundance was reduced to one-half in heterozygotes and was undetectable in Sumo-1(-/-) mice, and SUMO-1-conjugated RanGAP1 was detectable in wild-type mouse embryo fibroblasts (MEFs) but not in Sumo-1(-/-) MEFs, indicating that gene targeting yielded Sumo-1-null mice. Sumo-1 mRNA is expressed in all tissues of wild-type mice, and its abundance is highest in the testis, brain, lungs, and spleen. Sumo-2 and Sumo-3 mRNAs are also expressed in all tissues, but their abundance was not upregulated in Sumo-1-null mice. The development and function of testis are normal in the absence of Sumo-1, and Sumo-1(-)(/)(-) mice of both sexes are viable and fertile. In contrast to a previous report (F. S. Alkuraya et al., Science 313:1751, 2006), we did not observe embryonic or early postnatal demise of Sumo-1-targeted mice; genotypes of embryos and 21-day-old mice were of predicted Mendelian ratios, and there was no defect in lip and palate development in Sumo-1(+/-) or Sumo-1(-/-) embryos. The ability of Sumo-1(-/-) MEFs to differentiate into adipocyte was not different from that of wild-type MEFs. Collectively, our results support the notion that most, if not all, SUMO-1 functions are compensated for in vivo by SUMO-2 and SUMO-3.
Figures
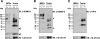
Similar articles
-
Loss of SUMO1 in mice affects RanGAP1 localization and formation of PML nuclear bodies, but is not lethal as it can be compensated by SUMO2 or SUMO3.J Cell Sci. 2008 Dec 15;121(Pt 24):4106-13. doi: 10.1242/jcs.038570. Epub 2008 Nov 25. J Cell Sci. 2008. PMID: 19033381
-
SUMO2 is essential while SUMO3 is dispensable for mouse embryonic development.EMBO Rep. 2014 Aug;15(8):878-85. doi: 10.15252/embr.201438534. Epub 2014 Jun 2. EMBO Rep. 2014. PMID: 24891386 Free PMC article.
-
SUMO paralogue-specific functions revealed through systematic analysis of human knockout cell lines and gene expression data.Mol Biol Cell. 2021 Sep 1;32(19):1849-1866. doi: 10.1091/mbc.E21-01-0031. Epub 2021 Jul 7. Mol Biol Cell. 2021. PMID: 34232706 Free PMC article.
-
DeSUMOylating enzymes--SENPs.IUBMB Life. 2008 Nov;60(11):734-42. doi: 10.1002/iub.113. IUBMB Life. 2008. PMID: 18666185 Review.
-
SUMO--nonclassical ubiquitin.Annu Rev Cell Dev Biol. 2000;16:591-626. doi: 10.1146/annurev.cellbio.16.1.591. Annu Rev Cell Dev Biol. 2000. PMID: 11031248 Review.
Cited by
-
Sumoylation of RORγt regulates TH17 differentiation and thymocyte development.Nat Commun. 2018 Nov 19;9(1):4870. doi: 10.1038/s41467-018-07203-z. Nat Commun. 2018. PMID: 30451821 Free PMC article.
-
A proteomic screen for nucleolar SUMO targets shows SUMOylation modulates the function of Nop5/Nop58.Mol Cell. 2010 Aug 27;39(4):618-31. doi: 10.1016/j.molcel.2010.07.025. Mol Cell. 2010. PMID: 20797632 Free PMC article.
-
Sumoylation in Physiology, Pathology and Therapy.Cells. 2022 Feb 26;11(5):814. doi: 10.3390/cells11050814. Cells. 2022. PMID: 35269436 Free PMC article. Review.
-
Signalling mechanisms and cellular functions of SUMO.Nat Rev Mol Cell Biol. 2022 Nov;23(11):715-731. doi: 10.1038/s41580-022-00500-y. Epub 2022 Jun 24. Nat Rev Mol Cell Biol. 2022. PMID: 35750927 Review.
-
Protection from isopeptidase-mediated deconjugation regulates paralog-selective sumoylation of RanGAP1.Mol Cell. 2009 Mar 13;33(5):570-80. doi: 10.1016/j.molcel.2009.02.008. Mol Cell. 2009. PMID: 19285941 Free PMC article.
References
-
- Alarcon-Vargas, D., and Z. Ronai. 2002. SUMO in cancer—wrestlers wanted. Cancer Biol. Ther. 1237-242. - PubMed
-
- Al-Khodairy, F., T. Enoch, I. M. Hagan, and A. M. Carr. 1995. The Schizosaccharomyces pombe hus5 gene encodes a ubiquitin conjugating enzyme required for normal mitosis. J. Cell Sci. 108475-486. - PubMed
-
- Alkuraya, F. S., I. Saadi, J. J. Lund, A. Turbe-Doan, C. C. Morton, and R. L. Maas. 2006. SUMO1 haploinsufficiency leads to cleft lip and palate. Science 3131751. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
