Cathepsin L functionally cleaves the severe acute respiratory syndrome coronavirus class I fusion protein upstream of rather than adjacent to the fusion peptide
- PMID: 18562523
- PMCID: PMC2519682
- DOI: 10.1128/JVI.00415-08
Cathepsin L functionally cleaves the severe acute respiratory syndrome coronavirus class I fusion protein upstream of rather than adjacent to the fusion peptide
Abstract
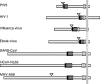
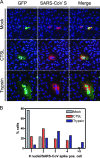
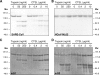
Unlike other class I viral fusion proteins, spike proteins on severe acute respiratory syndrome coronavirus virions are uncleaved. As we and others have demonstrated, infection by this virus depends on cathepsin proteases present in endosomal compartments of the target cell, suggesting that the spike protein acquires its fusion competence by cleavage during cell entry rather than during virion biogenesis. Here we demonstrate that cathepsin L indeed activates the membrane fusion function of the spike protein. Moreover, cleavage was mapped to the same region where, in coronaviruses carrying furin-activated spikes, the receptor binding subunit of the protein is separated from the membrane-anchored fusion subunit.
Figures

Similar articles
-
Endosomal proteolysis by cathepsins is necessary for murine coronavirus mouse hepatitis virus type 2 spike-mediated entry.J Virol. 2006 Jun;80(12):5768-76. doi: 10.1128/JVI.00442-06. J Virol. 2006. PMID: 16731916 Free PMC article.
-
Proteolysis of SARS-associated coronavirus spike glycoprotein.Adv Exp Med Biol. 2006;581:235-40. doi: 10.1007/978-0-387-33012-9_39. Adv Exp Med Biol. 2006. PMID: 17037535 Free PMC article. No abstract available.
-
SARS coronavirus, but not human coronavirus NL63, utilizes cathepsin L to infect ACE2-expressing cells.J Biol Chem. 2006 Feb 10;281(6):3198-203. doi: 10.1074/jbc.M508381200. Epub 2005 Dec 8. J Biol Chem. 2006. PMID: 16339146 Free PMC article.
-
A mature and fusogenic form of the Nipah virus fusion protein requires proteolytic processing by cathepsin L.Virology. 2006 Mar 15;346(2):251-7. doi: 10.1016/j.virol.2006.01.007. Epub 2006 Feb 7. Virology. 2006. PMID: 16460775 Free PMC article.
-
Unique neuronal functions of cathepsin L and cathepsin B in secretory vesicles: biosynthesis of peptides in neurotransmission and neurodegenerative disease.Biol Chem. 2006 Oct-Nov;387(10-11):1429-39. doi: 10.1515/BC.2006.179. Biol Chem. 2006. PMID: 17081116 Review.
Cited by
-
Potential Effects of Hyperglycemia on SARS-CoV-2 Entry Mechanisms in Pancreatic Beta Cells.Viruses. 2024 Aug 2;16(8):1243. doi: 10.3390/v16081243. Viruses. 2024. PMID: 39205219 Free PMC article. Review.
-
High-throughput screening identifies broad-spectrum Coronavirus entry inhibitors.iScience. 2024 May 17;27(6):110019. doi: 10.1016/j.isci.2024.110019. eCollection 2024 Jun 21. iScience. 2024. PMID: 38883823 Free PMC article.
-
The Effects of Chloroquine and Hydroxychloroquine on ACE2-Related Coronavirus Pathology and the Cardiovascular System: An Evidence-Based Review.Function (Oxf). 2020 Jul 27;1(2):zqaa012. doi: 10.1093/function/zqaa012. eCollection 2020. Function (Oxf). 2020. PMID: 38626250 Free PMC article.
-
Natural products as a source of Coronavirus entry inhibitors.Front Cell Infect Microbiol. 2024 Feb 21;14:1353971. doi: 10.3389/fcimb.2024.1353971. eCollection 2024. Front Cell Infect Microbiol. 2024. PMID: 38449827 Free PMC article. Review.
-
Genome-wide bioinformatics analysis of human protease capacity for proteolytic cleavage of the SARS-CoV-2 spike glycoprotein.Microbiol Spectr. 2024 Feb 6;12(2):e0353023. doi: 10.1128/spectrum.03530-23. Epub 2024 Jan 8. Microbiol Spectr. 2024. PMID: 38189333 Free PMC article.
References
-
- Bosch, B. J., B. E. Martina, R. Van Der Zee, J. Lepault, B. J. Haijema, C. Versluis, A. J. Heck, R. De Groot, A. D. Osterhaus, and P. J. Rottier. 2004. Severe acute respiratory syndrome coronavirus (SARS-CoV) infection inhibition using spike protein heptad repeat-derived peptides. Proc. Natl. Acad. Sci. USA 1018455-8460. - PMC - PubMed
-
- Calder, L. J., L. Gonzalez-Reyes, B. Garcia-Barreno, S. A. Wharton, J. J. Skehel, D. C. Wiley, and J. A. Melero. 2000. Electron microscopy of the human respiratory syncytial virus fusion protein and complexes that it forms with monoclonal antibodies. Virology 271122-131. - PubMed
-
- Chambers, P., C. R. Pringle, and A. J. Easton. 1990. Heptad repeat sequences are located adjacent to hydrophobic regions in several types of virus fusion glycoproteins. J. Gen. Virol. 713075-3080. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

