Maternal and zygotic Dnmt1 are necessary and sufficient for the maintenance of DNA methylation imprints during preimplantation development
- PMID: 18559477
- PMCID: PMC2428059
- DOI: 10.1101/gad.1667008
Maternal and zygotic Dnmt1 are necessary and sufficient for the maintenance of DNA methylation imprints during preimplantation development
Abstract
Parental origin-specific DNA methylation regulates the monoallelic expression of the mammalian imprinted genes. The methylation marks or imprints are established in the parental germline and maintained throughout embryonic development. However, it is unclear how the methylation imprints are maintained through extensive demethylation in cleavage-stage preimplantation embryos. Previous reports suggested that DNA methyltransferase(s) other than Dnmt1 is involved in the maintenance of the imprints during cleavage. Here we demonstrate, by using conditional knockout mice, that the other known DNA methyltransferases Dnmt3a and Dnmt3b are dispensable for the maintenance of the methylation marks at most imprinted loci. We further demonstrate that a lack of both maternal and zygotic Dnmt1 results in complete demethylation of all imprinted loci examined in blastocysts. Consistent with these results we find that zygotic Dnmt1 is expressed in the preimplantation embryo. Thus, contrary to the previous reports, Dnmt1 alone is sufficient to maintain the methylation marks of the imprinted genes.
Figures
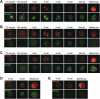

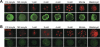

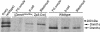
Comment in
-
Safeguarding parental identity: Dnmt1 maintains imprints during epigenetic reprogramming in early embryogenesis.Genes Dev. 2008 Jun 15;22(12):1567-71. doi: 10.1101/gad.1690508. Genes Dev. 2008. PMID: 18559472 Free PMC article.
Similar articles
-
Unfaithful maintenance of methylation imprints due to loss of maternal nuclear Dnmt1 during somatic cell nuclear transfer.PLoS One. 2011;6(5):e20154. doi: 10.1371/journal.pone.0020154. Epub 2011 May 20. PLoS One. 2011. PMID: 21625467 Free PMC article.
-
Safeguarding parental identity: Dnmt1 maintains imprints during epigenetic reprogramming in early embryogenesis.Genes Dev. 2008 Jun 15;22(12):1567-71. doi: 10.1101/gad.1690508. Genes Dev. 2008. PMID: 18559472 Free PMC article.
-
Identification of a region of the DNMT1 methyltransferase that regulates the maintenance of genomic imprints.Proc Natl Acad Sci U S A. 2009 Dec 8;106(49):20806-11. doi: 10.1073/pnas.0905668106. Epub 2009 Nov 18. Proc Natl Acad Sci U S A. 2009. PMID: 19923434 Free PMC article.
-
Maternal control of genomic imprint maintenance.Reprod Biomed Online. 2013 Dec;27(6):629-36. doi: 10.1016/j.rbmo.2013.06.004. Epub 2013 Jun 20. Reprod Biomed Online. 2013. PMID: 24125946 Review.
-
New insights into establishment and maintenance of DNA methylation imprints in mammals.Philos Trans R Soc Lond B Biol Sci. 2013 Jan 5;368(1609):20110336. doi: 10.1098/rstb.2011.0336. Philos Trans R Soc Lond B Biol Sci. 2013. PMID: 23166397 Free PMC article. Review.
Cited by
-
Pramel15 facilitates zygotic nuclear DNMT1 degradation and DNA demethylation.Nat Commun. 2024 Aug 25;15(1):7310. doi: 10.1038/s41467-024-51614-0. Nat Commun. 2024. PMID: 39181896 Free PMC article.
-
Assisted reproduction treatment and epigenetic inheritance.Hum Reprod Update. 2012 Mar-Apr;18(2):171-97. doi: 10.1093/humupd/dmr047. Epub 2012 Jan 19. Hum Reprod Update. 2012. PMID: 22267841 Free PMC article. Review.
-
Epigenetic mechanisms in heart development and disease.Drug Discov Today. 2015 Jul;20(7):799-811. doi: 10.1016/j.drudis.2014.12.018. Epub 2015 Jan 6. Drug Discov Today. 2015. PMID: 25572405 Free PMC article. Review.
-
DNA methyltransferase 1o functions during preimplantation development to preclude a profound level of epigenetic variation.Dev Biol. 2008 Dec 1;324(1):139-50. doi: 10.1016/j.ydbio.2008.09.015. Epub 2008 Sep 25. Dev Biol. 2008. PMID: 18845137 Free PMC article.
-
Developmental Functions of the Dynamic DNA Methylome and Hydroxymethylome in the Mouse and Zebrafish: Similarities and Differences.Front Cell Dev Biol. 2018 Mar 20;6:27. doi: 10.3389/fcell.2018.00027. eCollection 2018. Front Cell Dev Biol. 2018. PMID: 29616219 Free PMC article. Review.
References
-
- Carlson L.L., Page A.W., Bestor T.H. Properties and localization of DNA methyltransferase in preimplantation mouse embryos: Implications for genomic imprinting. Genes & Dev. 1992;6:2536–2541. - PubMed
-
- de Vries W.N., Binns L.T., Fancher K.S., Dean J., Moore R., Kemler R., Knowles B.B. Expression of Cre recombinase in mouse oocytes: A means to study maternal effect genes. Genesis. 2000;26:110–112. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
